Multiple Click-Selective tRNA Synthetases Expand Mammalian Cell-Specific Proteomics
- PMID: 29775058
- PMCID: PMC6598694
- DOI: 10.1021/jacs.8b03074
Multiple Click-Selective tRNA Synthetases Expand Mammalian Cell-Specific Proteomics
Abstract
Bioorthogonal tools enable cell-type-specific proteomics, a prerequisite to understanding biological processes in multicellular organisms. Here we report two engineered aminoacyl-tRNA synthetases for mammalian bioorthogonal labeling: a tyrosyl ( ScTyrY43G) and a phenylalanyl ( MmPheT413G) tRNA synthetase that incorporate azide-bearing noncanonical amino acids specifically into the nascent proteomes of host cells. Azide-labeled proteins are chemoselectively tagged via azide-alkyne cycloadditions with fluorophores for imaging or affinity resins for mass spectrometric characterization. Both mutant synthetases label human, hamster, and mouse cell line proteins and selectively activate their azido-bearing amino acids over 10-fold above the canonical. ScTyrY43G and MmPheT413G label overlapping but distinct proteomes in human cell lines, with broader proteome coverage upon their coexpression. In mice, ScTyrY43G and MmPheT413G label the melanoma tumor proteome and plasma secretome. This work furnishes new tools for mammalian residue-specific bioorthogonal chemistry, and enables more robust and comprehensive cell-type-specific proteomics in live mammals.
Figures
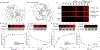
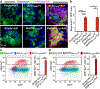

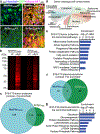
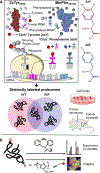
References
-
- Conboy IM; Conboy MJ; Wagers AJ; Girma ER; Weissman IL; Rando TA Nature 2005, 433 (7027), 760–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

