p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis
- PMID: 29775581
- PMCID: PMC6029451
- DOI: 10.1016/j.molcel.2018.04.011
p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis
Erratum in
-
p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis.Mol Cell. 2018 Jun 7;70(5):984. doi: 10.1016/j.molcel.2018.05.035. Epub 2018 Jun 7. Mol Cell. 2018. PMID: 29883613 Free PMC article. No abstract available.
Abstract
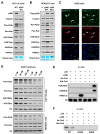
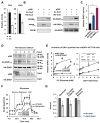
Lysine 2-hydroxyisobutyrylation (Khib) is an evolutionarily conserved and widespread histone mark like lysine acetylation (Kac). Here we report that p300 functions as a lysine 2-hyroxyisobutyryltransferase to regulate glycolysis in response to nutritional cues. We discovered that p300 differentially regulates Khib and Kac on distinct lysine sites, with only 6 of the 149 p300-targeted Khib sites overlapping with the 693 p300-targeted Kac sites. We demonstrate that diverse cellular proteins, particularly glycolytic enzymes, are targeted by p300 for Khib, but not for Kac. Specifically, deletion of p300 significantly reduces Khib levels on several p300-dependent, Khib-specific sites on key glycolytic enzymes including ENO1, decreasing their catalytic activities. Consequently, p300-deficient cells have impaired glycolysis and are hypersensitive to glucose-depletion-induced cell death. Our study reveals an p300-catalyzed, Khib-specific molecular mechanism that regulates cellular glucose metabolism and further indicate that p300 has an intrinsic ability to select short-chain acyl-CoA-dependent protein substrates.
Keywords: EP300; cell survival; glycolysis; lysine 2-hydroxyisobutyrylation.
Published by Elsevier Inc.
Conflict of interest statement
Y. Z. is on the science advisory board of PTM Biolabs.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

