Targeting cancer's metabolic co-dependencies: A landscape shaped by genotype and tissue context
- PMID: 29775654
- PMCID: PMC6193564
- DOI: 10.1016/j.bbcan.2018.05.002
Targeting cancer's metabolic co-dependencies: A landscape shaped by genotype and tissue context
Abstract
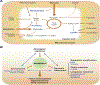
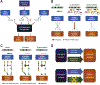
Tumors cells reprogram their metabolism to fuel rapid growth. The ability to trace nutrient fluxes in the context of specific alterations has provided new mechanistic insight into the process of oncogenic transformation. A broad array of complementary genetic, epigenetic, transcriptional and translational mechanisms has been identified, revealing a metabolic landscape of cancer. However, cancer metabolism is not a static or uniform process, including within a single tumor. Tumor cells adapt to changing environmental conditions, profoundly shaping the enzymatic dependencies of individual cells. The underlying molecular mechanisms of adaptation, and the specific interactions between tumor genotype, oncogenic signaling, and tissue/biochemical context, remain incompletely understood. In this review, we examine dynamic aspects of how metabolic dependencies develop in cancer, shaped both by genotype and biochemical environment, and review how these interlaced processes generate targetable metabolic vulnerabilities. This article is part of a Special Issue entitled: Cancer Metabolism edited by Dr. Chi Van Dang.
Keywords: Cancer metabolism; Heterogeneity; Metabolic co-dependency; Oncogenic signaling; Tissue context; ecDNA.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures

References
-
- Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP, Cancer metabolism: a therapeutic perspective, Nature reviews. Clinical oncology, 14 (2017) 11-31. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

