Defining Host Responses during Systemic Bacterial Infection through Construction of a Murine Organ Proteome Atlas
- PMID: 29778837
- PMCID: PMC7868092
- DOI: 10.1016/j.cels.2018.04.010
Defining Host Responses during Systemic Bacterial Infection through Construction of a Murine Organ Proteome Atlas
Abstract
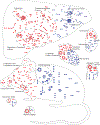
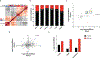
Group A Streptococcus (GAS) remains one of the top 10 deadliest human pathogens worldwide despite its sensitivity to penicillin. Although the most common GAS infection is pharyngitis (strep throat), it also causes life-threatening systemic infections. A series of complex networks between host and pathogen drive invasive infections, which have not been comprehensively mapped. Attempting to map these interactions, we examined organ-level protein dynamics using a mouse model of systemic GAS infection. We quantified over 11,000 proteins, defining organ-specific markers for all analyzed tissues. From this analysis, an atlas of dynamically regulated proteins and pathways was constructed. Through statistical methods, we narrowed organ-specific markers of infection to 34 from the defined atlas. We show these markers are trackable in blood of infected mice, and a subset has been observed in plasma samples from GAS-infected clinical patients. This proteomics-based strategy provides insight into host defense responses, establishes potentially useful targets for therapeutic intervention, and presents biomarkers for determining affected organs during bacterial infection.
Keywords: Orbitrap; S. pyogenes; Tandem Mass Tag; group A Streptococcus; multiplexed proteomics; systemic infection.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
All authors declare no competing interests.
Figures

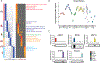
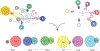

Comment in
-
A Host Proteome Atlas of Streptococcus pyogenes Infection.Cell Syst. 2018 May 23;6(5):536-538. doi: 10.1016/j.cels.2018.05.003. Cell Syst. 2018. PMID: 29792824
Similar articles
-
Differential compartmentalization of Streptococcus pyogenes virulence factors and host protein binding properties as a mechanism for host adaptation.Int J Med Microbiol. 2016 Nov;306(7):504-516. doi: 10.1016/j.ijmm.2016.06.007. Epub 2016 Jun 29. Int J Med Microbiol. 2016. PMID: 27423808
-
Targeted Proteomics and Absolute Protein Quantification for the Construction of a Stoichiometric Host-Pathogen Surface Density Model.Mol Cell Proteomics. 2017 Apr;16(4 suppl 1):S29-S41. doi: 10.1074/mcp.M116.063966. Epub 2017 Feb 9. Mol Cell Proteomics. 2017. PMID: 28183813 Free PMC article.
-
A quantitative Streptococcus pyogenes-human protein-protein interaction map reveals localization of opsonizing antibodies.Nat Commun. 2019 Jun 21;10(1):2727. doi: 10.1038/s41467-019-10583-5. Nat Commun. 2019. PMID: 31227708 Free PMC article.
-
Group A streptococcal pharyngitis: Immune responses involved in bacterial clearance and GAS-associated immunopathologies.J Leukoc Biol. 2018 Feb;103(2):193-213. doi: 10.1189/jlb.4MR0617-227RR. Epub 2017 Dec 29. J Leukoc Biol. 2018. PMID: 28951419 Review.
-
Mechanisms of group A Streptococcus resistance to reactive oxygen species.FEMS Microbiol Rev. 2015 Jul;39(4):488-508. doi: 10.1093/femsre/fuu009. Epub 2015 Feb 10. FEMS Microbiol Rev. 2015. PMID: 25670736 Free PMC article. Review.
Cited by
-
A Broad Spectrum Antiparasitic Activity of Organotin (IV) Derivatives and Its Untargeted Proteomic Profiling Using Leishmania donovani.Pathogens. 2022 Nov 26;11(12):1424. doi: 10.3390/pathogens11121424. Pathogens. 2022. PMID: 36558759 Free PMC article.
-
Functional and Proteomic Analysis of Streptococcus pyogenes Virulence Upon Loss of Its Native Cas9 Nuclease.Front Microbiol. 2019 Aug 22;10:1967. doi: 10.3389/fmicb.2019.01967. eCollection 2019. Front Microbiol. 2019. PMID: 31507572 Free PMC article.
-
EGFR is required for Wnt9a-Fzd9b signalling specificity in haematopoietic stem cells.Nat Cell Biol. 2019 Jun;21(6):721-730. doi: 10.1038/s41556-019-0330-5. Epub 2019 May 20. Nat Cell Biol. 2019. PMID: 31110287 Free PMC article.
-
The intricate pathogenicity of Group A Streptococcus: A comprehensive update.Virulence. 2024 Dec;15(1):2412745. doi: 10.1080/21505594.2024.2412745. Epub 2024 Nov 5. Virulence. 2024. PMID: 39370779 Free PMC article. Review.
-
Vascular Proteome Responses Precede Organ Dysfunction in a Murine Model of Staphylococcus aureus Bacteremia.mSystems. 2022 Aug 30;7(4):e0039522. doi: 10.1128/msystems.00395-22. Epub 2022 Aug 1. mSystems. 2022. PMID: 35913192 Free PMC article.
References
-
- AMMANN P, FEHR T, MINDER EI, GUNTER C & BERTEL O (2001). Elevation of troponin I in sepsis and septic shock. Intensive Care Med, 27, 965–9. - PubMed
-
- ASHBAUGH CD, MOSER TJ, SHEARER MH, WHITE GL, KENNEDY RC & WESSELS MR (2000). Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cell Microbiol, 2, 283–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

