Non-canonical Phototransduction Mediates Synchronization of the Drosophila melanogaster Circadian Clock and Retinal Light Responses
- PMID: 29779871
- PMCID: PMC5988559
- DOI: 10.1016/j.cub.2018.04.016
Non-canonical Phototransduction Mediates Synchronization of the Drosophila melanogaster Circadian Clock and Retinal Light Responses
Abstract
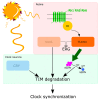
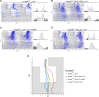
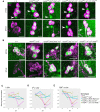
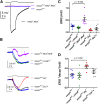
The daily light-dark cycles represent a key signal for synchronizing circadian clocks. Both insects and mammals possess dedicated "circadian" photoreceptors but also utilize the visual system for clock resetting. In Drosophila, circadian clock resetting is achieved by the blue-light photoreceptor cryptochrome (CRY), which is expressed within subsets of the brain clock neurons. In addition, rhodopsin-expressing photoreceptor cells contribute to light synchronization. Light resets the molecular clock by CRY-dependent degradation of the clock protein Timeless (TIM), although in specific subsets of key circadian pacemaker neurons, including the small ventral lateral neurons (s-LNvs), TIM and Period (PER) oscillations can be synchronized by light independent of CRY and canonical visual Rhodopsin phototransduction. Here, we show that at least three of the seven Drosophila rhodopsins can utilize an alternative transduction mechanism involving the same α-subunit of the heterotrimeric G protein operating in canonical visual phototransduction (Gq). Surprisingly, in mutants lacking the canonical phospholipase C-β (PLC-β) encoded by the no receptor potential A (norpA) gene, we uncovered a novel transduction pathway using a different PLC-β encoded by the Plc21C gene. This novel pathway is important for behavioral clock resetting to semi-natural light-dark cycles and mediates light-dependent molecular synchronization within the s-LNv clock neurons. The same pathway appears to be responsible for norpA-independent light responses in the compound eye. We show that Rhodopsin 5 (Rh5) and Rh6, present in the R8 subset of retinal photoreceptor cells, drive both the long-term circadian and rapid light responses in the eye.
Keywords: circadian clock; cryptochrome; period; phospholipase C; phototransduction; rhodopsin; timeless.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures

References
-
- Emery P., So W.V., Kaneko M., Hall J.C., Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. - PubMed
-
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., Kay S.A., Rosbash M., Hall J.C. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. - PubMed
-
- Busza A., Emery-Le M., Rosbash M., Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

