Automated typing of red blood cell and platelet antigens: a whole-genome sequencing study
- PMID: 29780001
- PMCID: PMC6438177
- DOI: 10.1016/S2352-3026(18)30053-X
Automated typing of red blood cell and platelet antigens: a whole-genome sequencing study
Abstract
Background: There are more than 300 known red blood cell (RBC) antigens and 33 platelet antigens that differ between individuals. Sensitisation to antigens is a serious complication that can occur in prenatal medicine and after blood transfusion, particularly for patients who require multiple transfusions. Although pre-transfusion compatibility testing largely relies on serological methods, reagents are not available for many antigens. Methods based on single-nucleotide polymorphism (SNP) arrays have been used, but typing for ABO and Rh-the most important blood groups-cannot be done with SNP typing alone. We aimed to develop a novel method based on whole-genome sequencing to identify RBC and platelet antigens.
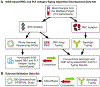
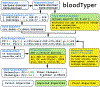
Methods: This whole-genome sequencing study is a subanalysis of data from patients in the whole-genome sequencing arm of the MedSeq Project randomised controlled trial (NCT01736566) with no measured patient outcomes. We created a database of molecular changes in RBC and platelet antigens and developed an automated antigen-typing algorithm based on whole-genome sequencing (bloodTyper). This algorithm was iteratively improved to address cis-trans haplotype ambiguities and homologous gene alignments. Whole-genome sequencing data from 110 MedSeq participants (30 × depth) were used to initially validate bloodTyper through comparison with conventional serology and SNP methods for typing of 38 RBC antigens in 12 blood-group systems and 22 human platelet antigens. bloodTyper was further validated with whole-genome sequencing data from 200 INTERVAL trial participants (15 × depth) with serological comparisons.
Findings: We iteratively improved bloodTyper by comparing its typing results with conventional serological and SNP typing in three rounds of testing. The initial whole-genome sequencing typing algorithm was 99·5% concordant across the first 20 MedSeq genomes. Addressing discordances led to development of an improved algorithm that was 99·8% concordant for the remaining 90 MedSeq genomes. Additional modifications led to the final algorithm, which was 99·2% concordant across 200 INTERVAL genomes (or 99·9% after adjustment for the lower depth of coverage).
Interpretation: By enabling more precise antigen-matching of patients with blood donors, antigen typing based on whole-genome sequencing provides a novel approach to improve transfusion outcomes with the potential to transform the practice of transfusion medicine.
Funding: National Human Genome Research Institute, Doris Duke Charitable Foundation, National Health Service Blood and Transplant, National Institute for Health Research, and Wellcome Trust.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures






References
-
- Hendrickson JE, Tormey CA, Shaz BH. Red blood cell alloimmunization mitigation strategies. Transfusion Medicine Reviews 2014; 28(3): 137–44. - PubMed
-
- Fung MK, Eder AF, Spitalnik SL, Westhoff CM, editors. Technical Manual, 19th edition. 19th ed: American Association of Blood Banks (AABB); 2017.
-
- Paccapelo C, Truglio F, Antonietta Villa M, Revelli N, Marconi M. HEA BeadChip technology in immunohematology. Immunohematology / American Red Cross 2015; 31(2): 81–90. - PubMed
-
- Hashmi G, Shariff T, Zhang Y, et al. Determination of 24 minor red blood cell antigens for more than 2000 blood donors by high-throughput DNA analysis. Transfusion 2007; 47(4): 736–47. - PubMed
-
- Stabentheiner S, Danzer M, Niklas N, et al. Overcoming methodical limits of standard RHD genotyping by next-generation sequencing. Vox Sanguinis 2011; 100(4): 381–8. - PubMed
MeSH terms
Substances
Grants and funding
- RF1 AG047866/AG/NIA NIH HHS/United States
- RG/13/13/30194/BHF_/British Heart Foundation/United Kingdom
- T32 HL007627/HL/NHLBI NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- U19 HD077671/HD/NICHD NIH HHS/United States
- R01 CA154517/CA/NCI NIH HHS/United States
- U01 HG006500/HG/NHGRI NIH HHS/United States
- P01 HL095489/HL/NHLBI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- MR/L003120/1/MRC_/Medical Research Council/United Kingdom
- P60 AR047782/AR/NIAMS NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- R03 HG008809/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

