Comparative Metatranscriptomics of Wheat Rhizosphere Microbiomes in Disease Suppressive and Non-suppressive Soils for Rhizoctonia solani AG8
- PMID: 29780371
- PMCID: PMC5945926
- DOI: 10.3389/fmicb.2018.00859
Comparative Metatranscriptomics of Wheat Rhizosphere Microbiomes in Disease Suppressive and Non-suppressive Soils for Rhizoctonia solani AG8
Abstract
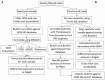
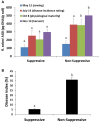
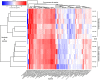
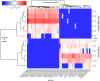
The soilborne fungus Rhizoctonia solani anastomosis group (AG) 8 is a major pathogen of grain crops resulting in substantial production losses. In the absence of resistant cultivars of wheat or barley, a sustainable and enduring method for disease control may lie in the enhancement of biological disease suppression. Evidence of effective biological control of R. solani AG8 through disease suppression has been well documented at our study site in Avon, South Australia. A comparative metatranscriptomic approach was applied to assess the taxonomic and functional characteristics of the rhizosphere microbiome of wheat plants grown in adjacent fields which are suppressive and non-suppressive to the plant pathogen R. solani AG8. Analysis of 12 rhizosphere metatranscriptomes (six per field) was undertaken using two bioinformatic approaches involving unassembled and assembled reads. Differential expression analysis showed the dominant taxa in the rhizosphere based on mRNA annotation were Arthrobacter spp. and Pseudomonas spp. for non-suppressive samples and Stenotrophomonas spp. and Buttiauxella spp. for the suppressive samples. The assembled metatranscriptome analysis identified more differentially expressed genes than the unassembled analysis in the comparison of suppressive and non-suppressive samples. Suppressive samples showed greater expression of a polyketide cyclase, a terpenoid biosynthesis backbone gene (dxs) and many cold shock proteins (csp). Non-suppressive samples were characterised by greater expression of antibiotic genes such as non-heme chloroperoxidase (cpo) which is involved in pyrrolnitrin synthesis, and phenazine biosynthesis family protein F (phzF) and its transcriptional activator protein (phzR). A large number of genes involved in detoxifying reactive oxygen species (ROS) and superoxide radicals (sod, cat, ahp, bcp, gpx1, trx) were also expressed in the non-suppressive rhizosphere samples most likely in response to the infection of wheat roots by R. solani AG8. Together these results provide new insight into microbial gene expression in the rhizosphere of wheat in soils suppressive and non-suppressive to R. solani AG8. The approach taken and the genes involved in these functions provide direction for future studies to determine more precisely the molecular interplay of plant-microbe-pathogen interactions with the ultimate goal of the development of management options that promote beneficial rhizosphere microflora to reduce R. solani AG8 infection of crops.
Keywords: Rhizoctonia root rot; differential gene expression; disease suppression; metatranscriptome assembly; microbiome; rhizosphere; soil; soilborne fungus.
Figures

References
-
- Alabouvette C. (1986). Fusarium-wilt suppressive soils from the Chateaurenard region: review of a 10-year study. Agronomie 6, 273–284. 10.1051/agro:19860307 - DOI
-
- Almasudy A. M., You M. P., Barbetti M. J. (2015). Influence of fungicidal seed treatments and soil type on severity of root disease caused by Rhizoctonia solani AG-8 on wheat. Crop Prot. 75, 40–45. 10.1016/j.cropro.2015.05.007 - DOI
-
- Anees M., Tronsmo A., Edel-Hermann V., Gautheron N., Faloya V., Steinberg C. (2010). Biotic changes in relation to local decrease in soil conduciveness to disease caused by Rhizoctonia solani. Eur. J. Plant Pathol. 126, 29–41. 10.1007/s10658-009-9517-0 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

