Counting charges on membrane-bound peptides
- PMID: 29780560
- PMCID: PMC5944241
- DOI: 10.1039/c8sc00804c
Counting charges on membrane-bound peptides
Abstract
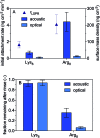
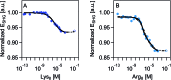
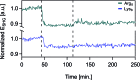
Quantifying the number of charges on peptides bound to interfaces requires reliable estimates of (i) surface coverage and (ii) surface charge, both of which are notoriously difficult parameters to obtain, especially at solid/water interfaces. Here, we report the thermodynamics and electrostatics governing the interactions of l-lysine and l-arginine octamers (Lys8 and Arg8) with supported lipid bilayers prepared from a 9 : 1 mixture of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dimyristoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (DMPG) from second harmonic generation (SHG) spectroscopy, quartz crystal microbalance with dissipation monitoring (QCM-D) and nanoplasmonic sensing (NPS) mass measurements, and atomistic simulations. The combined SHG/QCM-D/NPS approach provides interfacial charge density estimates from mean field theory for the attached peptides that are smaller by a factor of approximately two (0.12 ± 0.03 C m-2 for Lys8 and 0.10 ± 0.02 C m-2 for Arg8) relative to poly-l-lysine and poly-l-arginine. These results, along with atomistic simulations, indicate that the surface charge density of the supported lipid bilayer is neutralized by the attached cationic peptides. Moreover, the number of charges associated with each attached peptide is commensurate with those found in solution; that is, Lys8 and Arg8 are fully ionized when attached to the bilayer. Computer simulations indicate Lys8 is more likely than Arg8 to "stand-up" on the surface, interacting with lipid headgroups through one or two sidechains while Arg8 is more likely to assume a "buried" conformation, interacting with the bilayer through up to six sidechains. Analysis of electrostatic potential and charge distribution from atomistic simulations suggests that the Gouy-Chapman model, which is widely used for mapping surface potential to surface charge, is semi-quantitatively valid; despite considerable orientational preference of interfacial water, the apparent dielectric constant for the interfacial solvent is about 30, due to the thermal fluctuation of the lipid-water interface.
Figures
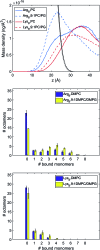

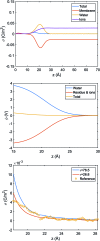
References
-
- Salay L. C., Petri D. F. S., Nakaie C. R., Schreier S. Biophys. Chem. 2015;207:128–134. - PubMed
-
- Wilhelm M. J., Gh M. S., Dai H. L. Biochemistry. 2015;54:4427–4430. - PubMed
-
- Nystrom L., Nordstrom R., Bramhill J., Saunders B. R., Alvarez-Asencio R., Rutland M. W., Malmsten M. Biomacromolecules. 2016;17:669–678. - PubMed
-
- Epand R. M., Epand R. F. Biochim. Biophys. Acta, Biomembr. 2009;1788:289–294. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

