Biomarker microRNAs for prostate cancer metastasis: screened with a network vulnerability analysis model
- PMID: 29784056
- PMCID: PMC5963164
- DOI: 10.1186/s12967-018-1506-7
Biomarker microRNAs for prostate cancer metastasis: screened with a network vulnerability analysis model
Abstract
Background: Prostate cancer (PCa) is a fatal malignant tumor among males in the world and the metastasis is a leading cause for PCa death. Biomarkers are therefore urgently needed to detect PCa metastatic signature at the early time. MicroRNAs are small non-coding RNAs with the potential to be biomarkers for disease prediction. In addition, computer-aided biomarker discovery is now becoming an attractive paradigm for precision diagnosis and prognosis of complex diseases.
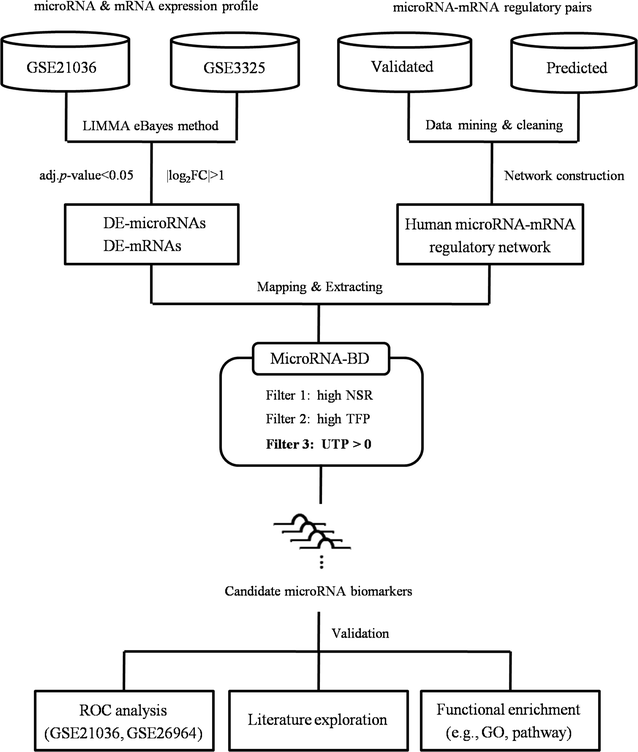
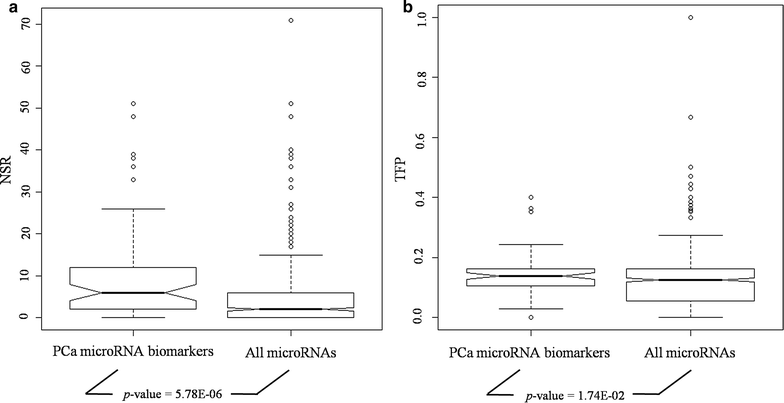
Methods: In this study, we identified key microRNAs as biomarkers for predicting PCa metastasis based on network vulnerability analysis. We first extracted microRNAs and mRNAs that were differentially expressed between primary PCa and metastatic PCa (MPCa) samples. Then we constructed the MPCa-specific microRNA-mRNA network and screened microRNA biomarkers by a novel bioinformatics model. The model emphasized the characterization of systems stability changes and the network vulnerability with three measurements, i.e. the structurally single-line regulation, the functional importance of microRNA targets and the percentage of transcription factor genes in microRNA unique targets.
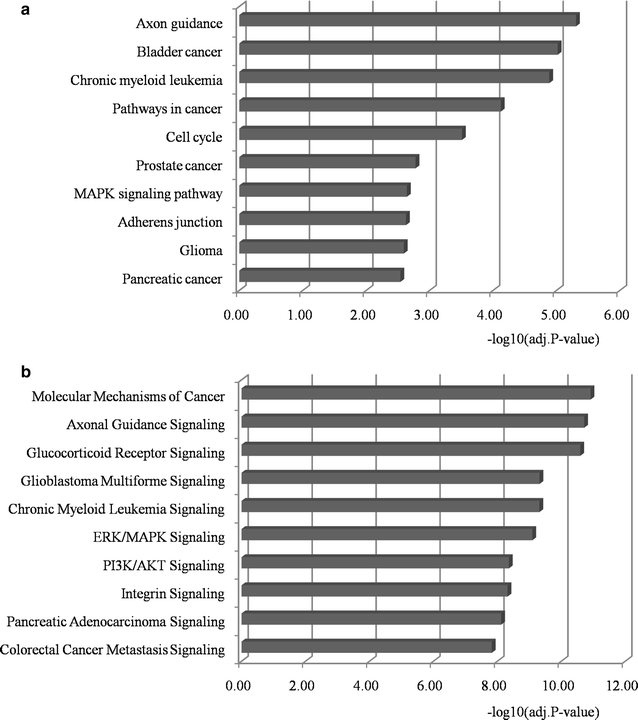
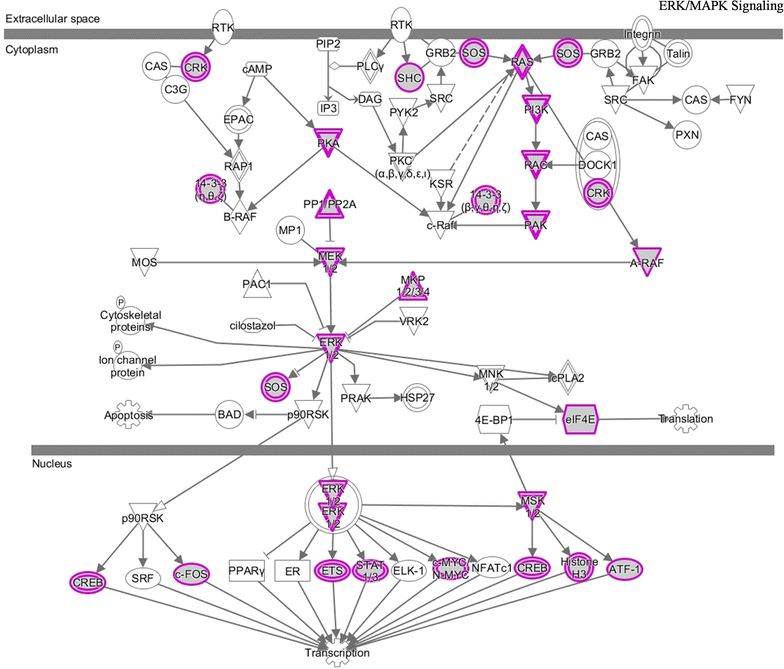
Results: With this model, we identified five microRNAs as putative biomarkers for PCa metastasis. Among them, miR-101-3p and miR-145-5p have been previously reported as biomarkers for PCa metastasis and the remaining three, i.e. miR-204-5p, miR-198 and miR-152, were screened as novel biomarkers for PCa metastasis. The results were further confirmed by the assessment of their predictive power and biological function analysis.
Conclusions: Five microRNAs were identified as candidate biomarkers for predicting PCa metastasis based on our network vulnerability analysis model. The prediction performance, literature exploration and functional enrichment analysis convinced our findings. This novel bioinformatics model could be applied to biomarker discovery for other complex diseases.
Keywords: Bioinformatics model; MicroRNA biomarkers; Network vulnerability analysis; Prostate cancer metastasis.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 31670851/National Natural Science Foundation of China/International
- 31470821/National Natural Science Foundation of China/International
- 91530320/National Natural Science Foundation of China/International
- 31770903/National Natural Science Foundation of China/International
- 2016YFC1306605/National Key Research and Development Program of China/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

