Protein-coding genes combined with long noncoding RNA as a novel transcriptome molecular staging model to predict the survival of patients with esophageal squamous cell carcinoma
- PMID: 29784063
- PMCID: PMC5993132
- DOI: 10.1186/s40880-018-0277-0
Protein-coding genes combined with long noncoding RNA as a novel transcriptome molecular staging model to predict the survival of patients with esophageal squamous cell carcinoma
Abstract
Background: Esophageal squamous cell carcinoma (ESCC) is the predominant subtype of esophageal carcinoma in China. This study was to develop a staging model to predict outcomes of patients with ESCC.
Methods: Using Cox regression analysis, principal component analysis (PCA), partitioning clustering, Kaplan-Meier analysis, receiver operating characteristic (ROC) curve analysis, and classification and regression tree (CART) analysis, we mined the Gene Expression Omnibus database to determine the expression profiles of genes in 179 patients with ESCC from GSE63624 and GSE63622 dataset.
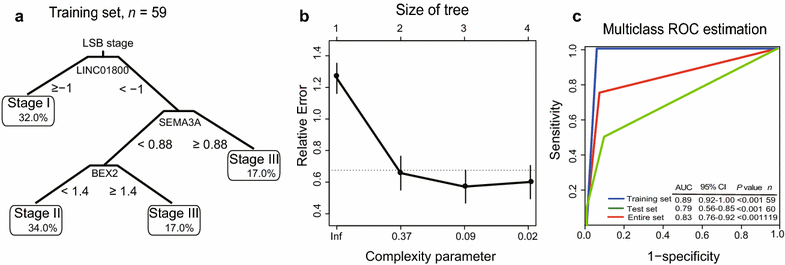
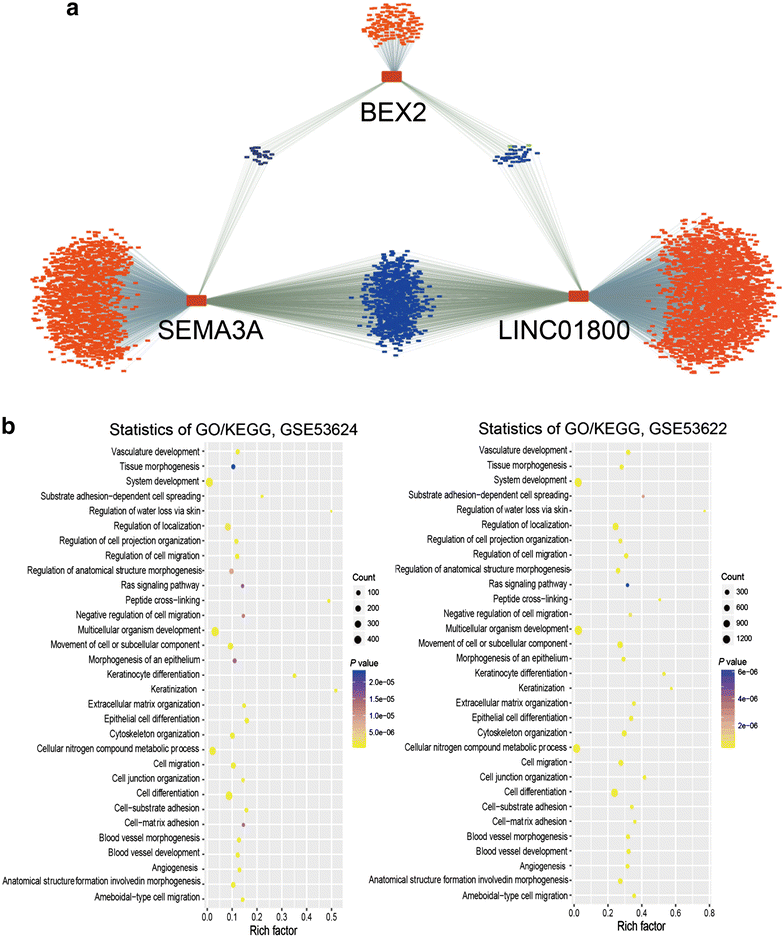
Results: Univariate cox regression analysis of the GSE63624 dataset revealed that 2404 protein-coding genes (PCGs) and 635 long non-coding RNAs (lncRNAs) were associated with the survival of patients with ESCC. PCA categorized these PCGs and lncRNAs into three principal components (PCs), which were used to cluster the patients into three groups. ROC analysis demonstrated that the predictive ability of PCG-lncRNA PCs when applied to new patients was better than that of the tumor-node-metastasis staging (area under ROC curve [AUC]: 0.69 vs. 0.65, P < 0.05). Accordingly, we constructed a molecular disaggregated model comprising one lncRNA and two PCGs, which we designated as the LSB staging model using CART analysis in the GSE63624 dataset. This LSB staging model classified the GSE63622 dataset of patients into three different groups, and its effectiveness was validated by analysis of another cohort of 105 patients.
Conclusions: The LSB staging model has clinical significance for the prognosis prediction of patients with ESCC and may serve as a three-gene staging microarray.
Keywords: Esophageal squamous cell carcinoma; Long non-coding RNA; Overall survival; Protein-coding gene; Staging model; Transcriptome.
Figures




References
-
- Takeno S, Noguchi T, Takahashi Y, Fumoto S, Shibata T, Kawahara K. Assessment of clinical outcome in patients with esophageal squamous cell carcinoma using TNM classification score and molecular biological classification. Ann Surg Oncol. 2007;14:1431–1438. doi: 10.1245/s10434-006-9286-3. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U1301227/the Natural Science Foundation of China-Guangdong Joint Fund/International
- U0932001/the Natural Science Foundation of China-Guangdong Joint Fund/International
- 81360331/the National Science Foundation of China/International
- 81472613/the National Science Foundation of China/International
- 81572341/the National Science Foundation of China/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

