Autophagy, Cell Viability, and Chemoresistance Are Regulated By miR-489 in Breast Cancer
- PMID: 29784669
- PMCID: PMC6843115
- DOI: 10.1158/1541-7786.MCR-17-0634
Autophagy, Cell Viability, and Chemoresistance Are Regulated By miR-489 in Breast Cancer
Abstract
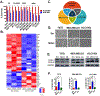
It is postulated that the complexity and heterogeneity in cancer may hinder most efforts that target a single pathway. Thus, discovery of novel therapeutic agents targeting multiple pathways, such as miRNAs, holds promise for future cancer therapy. One such miRNA, miR-489, is downregulated in a majority of breast cancer cells and several drug-resistant breast cancer cell lines, but its role and underlying mechanism for tumor suppression and drug resistance needs further investigation. The current study identifies autophagy as a novel pathway targeted by miR-489 and reports Unc-51 like autophagy activating kinase 1 (ULK1) and lysosomal protein transmembrane 4 beta (LAPTM4B) to be direct targets of miR-489. Furthermore, the data demonstrate autophagy inhibition and LAPTM4B downregulation as a major mechanism responsible for miR-489-mediated doxorubicin sensitization. Finally, miR-489 and LAPTM4B levels were inversely correlated in human tumor clinical specimens, and more importantly, miR-489 expression levels predict overall survival in patients with 8q22 amplification (the region in which LAPTM4B resides).Implications: These findings expand the understanding of miR-489-mediated tumor suppression and chemosensitization in and suggest a strategy for using miR-489 as a therapeutic sensitizer in a defined subgroup of resistant breast cancer patients. Mol Cancer Res; 16(9); 1348-60. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
Disclosure of potential conflicts of interest
The authors have no potential conflict of interest to disclosure.
Figures


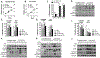


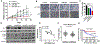
References
-
- Esparza-Lopez J, Escobar-Arriaga E, Soto-Germes S, Ibarra-Sanchez MJ. Breast Cancer Intra-Tumor Heterogeneity: One Tumor, Different Entities. Revista de investigacion clinica; organo del Hospital de Enfermedades de la Nutricion 2017;69(2):66–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

