Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis
- PMID: 29784811
- PMCID: PMC6003329
- DOI: 10.1073/pnas.1803440115
Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis
Abstract
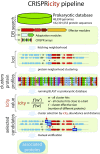
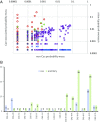
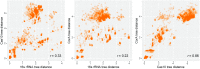
The CRISPR-Cas systems of bacterial and archaeal adaptive immunity consist of direct repeat arrays separated by unique spacers and multiple CRISPR-associated (cas) genes encoding proteins that mediate all stages of the CRISPR response. In addition to the relatively small set of core cas genes that are typically present in all CRISPR-Cas systems of a given (sub)type and are essential for the defense function, numerous genes occur in CRISPR-cas loci only sporadically. Some of these have been shown to perform various ancillary roles in CRISPR response, but the functional relevance of most remains unknown. We developed a computational strategy for systematically detecting genes that are likely to be functionally linked to CRISPR-Cas. The approach is based on a "CRISPRicity" metric that measures the strength of CRISPR association for all protein-coding genes from sequenced bacterial and archaeal genomes. Uncharacterized genes with CRISPRicity values comparable to those of cas genes are considered candidate CRISPR-linked genes. We describe additional criteria to predict functionally relevance for genes in the candidate set and identify 79 genes as strong candidates for functional association with CRISPR-Cas systems. A substantial majority of these CRISPR-linked genes reside in type III CRISPR-cas loci, which implies exceptional functional versatility of type III systems. Numerous candidate CRISPR-linked genes encode integral membrane proteins suggestive of tight membrane association of CRISPR-Cas systems, whereas many others encode proteins implicated in various signal transduction pathways. These predictions provide ample material for improving annotation of CRISPR-cas loci and experimental characterization of previously unsuspected aspects of CRISPR-Cas system functionality.
Keywords: CRISPR-Cas; computational genomics; gene neighborhoods; membrane proteins; signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. - PubMed
-
- Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. - PubMed
-
- Mohanraju P, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. - PubMed
-
- Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2:17092. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

