VirB8 homolog TraE from plasmid pKM101 forms a hexameric ring structure and interacts with the VirB6 homolog TraD
- PMID: 29784815
- PMCID: PMC6003364
- DOI: 10.1073/pnas.1802501115
VirB8 homolog TraE from plasmid pKM101 forms a hexameric ring structure and interacts with the VirB6 homolog TraD
Abstract
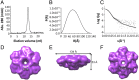
Type IV secretion systems (T4SSs) are multiprotein assemblies that translocate macromolecules across the cell envelope of bacteria. X-ray crystallographic and electron microscopy (EM) analyses have increasingly provided structural information on individual T4SS components and on the entire complex. As of now, relatively little information has been available on the exact localization of the inner membrane-bound T4SS components, notably the mostly periplasmic VirB8 protein and the very hydrophobic VirB6 protein. We show here that the membrane-bound, full-length version of the VirB8 homolog TraE from the plasmid pKM101 secretion system forms a high-molecular-mass complex that is distinct from the previously characterized periplasmic portion of the protein that forms dimers. Full-length TraE was extracted from the membranes with detergents, and analysis by size-exclusion chromatography, cross-linking, and size exclusion chromatography (SEC) multiangle light scattering (MALS) shows that it forms a high-molecular-mass complex. EM and small-angle X-ray scattering (SAXS) analysis demonstrate that full-length TraE forms a hexameric complex with a central pore. We also overproduced and purified the VirB6 homolog TraD and show by cross-linking, SEC, and EM that it binds to TraE. Our results suggest that TraE and TraD interact at the substrate translocation pore of the secretion system.
Keywords: VirB6-like; VirB8-like; conjugation; plasmid; type IV secretion.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

