Xenoprotein engineering via synthetic libraries
- PMID: 29784819
- PMCID: PMC6003312
- DOI: 10.1073/pnas.1722633115
Xenoprotein engineering via synthetic libraries
Abstract
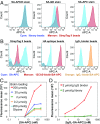
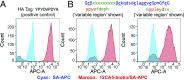
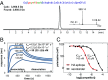
Chemical methods have enabled the total synthesis of protein molecules of ever-increasing size and complexity. However, methods to engineer synthetic proteins comprising noncanonical amino acids have not kept pace, even though this capability would be a distinct advantage of the total synthesis approach to protein science. In this work, we report a platform for protein engineering based on the screening of synthetic one-bead one-compound protein libraries. Screening throughput approaching that of cell surface display was achieved by a combination of magnetic bead enrichment, flow cytometry analysis of on-bead screens, and high-throughput MS/MS-based sequencing of identified active compounds. Direct screening of a synthetic protein library by these methods resulted in the de novo discovery of mirror-image miniprotein-based binders to a ∼150-kDa protein target, a task that would be difficult or impossible by other means.
Keywords: D-protein; flow cytometry; mirror-image miniprotein; protein engineering; xenoprotein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2004;44:34–66. - PubMed
-
- Jackson JC, Duffy SP, Hess KR, Mehl RA. Improving nature’s enzyme active site with genetically encoded unnatural amino acids. J Am Chem Soc. 2006;128:11124–11127. - PubMed
-
- Ugwumba IN, et al. Improving a natural enzyme activity through incorporation of unnatural amino acids. J Am Chem Soc. 2011;133:326–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

