Biological species in the viral world
- PMID: 29784828
- PMCID: PMC6003344
- DOI: 10.1073/pnas.1717593115
Biological species in the viral world
Abstract
Due to their dependence on cellular organisms for metabolism and replication, viruses are typically named and assigned to species according to their genome structure and the original host that they infect. But because viruses often infect multiple hosts and the numbers of distinct lineages within a host can be vast, their delineation into species is often dictated by arbitrary sequence thresholds, which are highly inconsistent across lineages. Here we apply an approach to determine the boundaries of viral species based on the detection of gene flow within populations, thereby defining viral species according to the biological species concept (BSC). Despite the potential for gene transfer between highly divergent genomes, viruses, like the cellular organisms they infect, assort into reproductively isolated groups and can be organized into biological species. This approach revealed that BSC-defined viral species are often congruent with the taxonomic partitioning based on shared gene contents and host tropism, and that bacteriophages can similarly be classified in biological species. These results open the possibility to use a single, universal definition of species that is applicable across cellular and acellular lifeforms.
Keywords: asexuality; biological species concept; gene flow; recombination; speciation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
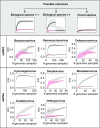
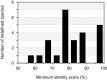

References
-
- Abedon ST, Calendar RL. The Bacteriophages. 2nd Ed. Oxford Univ Press; Oxford: 2005. p. 768.
-
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus Taxonomy: VIIIth Report of the International Committee on Tanoxomy of Viruses. Elsevier Academic; London: 2005.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

