Genome Variation and Molecular Epidemiology of Salmonella enterica Serovar Typhimurium Pathovariants
- PMID: 29784861
- PMCID: PMC6056856
- DOI: 10.1128/IAI.00079-18
Genome Variation and Molecular Epidemiology of Salmonella enterica Serovar Typhimurium Pathovariants
Abstract
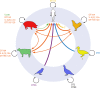
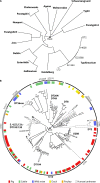
Salmonella enterica serovar Typhimurium is one of approximately 2,500 distinct serovars of the genus Salmonella but is exceptional in its wide distribution in the environment, livestock, and wild animals. S Typhimurium causes a large proportion of nontyphoidal Salmonella (NTS) infections, accounting for a quarter of infections, second only to S. enterica serovar Enteritidis in incidence. S Typhimurium was once considered the archetypal broad-host-range Salmonella serovar due to its wide distribution in livestock and wild animals, and much of what we know of the interaction of Salmonella with the host comes from research using a small number of laboratory strains of the serovar (LT2, SL1344, and ATCC 14028). But it has become clear that these strains do not reflect the genotypic or phenotypic diversity of S Typhimurium. Here, we review the epidemiological record of S Typhimurium and studies of the host-pathogen interactions of diverse strains of S Typhimurium. We present the concept of distinct pathovariants of S Typhimurium that exhibit diversity of host range, distribution in the environment, pathogenicity, and risk to food safety. We review recent evidence from whole-genome sequencing that has revealed the extent of genomic diversity of S Typhimurium pathovariants, the genomic basis of differences in the level of risk to human and animal health, and the molecular epidemiology of prominent strains. An improved understanding of the impact of genome variation of bacterial pathogens on pathogen-host and pathogen-environment interactions has the potential to improve quantitative risk assessment and reveal how new pathogens evolve.
Keywords: Salmonella; Typhimurium; epidemiology; genomics; host adaptation; phylogenetics; virulence.
© Crown copyright 2018.
Figures
References
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies . 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi:10.1086/650733. - DOI - PubMed
-
- Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull World Health Organ 82:346–353. http://europepmc.org/abstract/MED/15298225. - PMC - PubMed
-
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi:10.1371/journal.pmed.1001921. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

