Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses
- PMID: 29784908
- PMCID: PMC6022810
- DOI: 10.1038/s41590-018-0121-3
Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses
Abstract
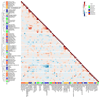
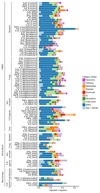
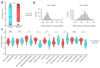
The immune response to pathogens varies substantially among people. Whereas both genetic and nongenetic factors contribute to interperson variation, their relative contributions and potential predictive power have remained largely unknown. By systematically correlating host factors in 534 healthy volunteers, including baseline immunological parameters and molecular profiles (genome, metabolome and gut microbiome), with cytokine production after stimulation with 20 pathogens, we identified distinct patterns of co-regulation. Among the 91 different cytokine-stimulus pairs, 11 categories of host factors together explained up to 67% of interindividual variation in cytokine production induced by stimulation. A computational model based on genetic data predicted the genetic component of stimulus-induced cytokine production (correlation 0.28-0.89), and nongenetic factors influenced cytokine production as well.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Netea MG, Wijmenga C, O’Neill LAJ. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol. 2012;13:535–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

