Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland
- PMID: 29784917
- PMCID: PMC6984964
- DOI: 10.1038/s41556-018-0108-1
Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland
Abstract
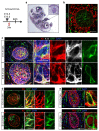
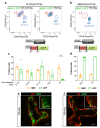
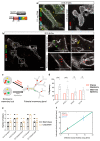
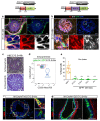
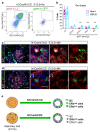
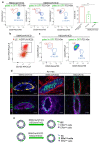
Recent lineage tracing studies have revealed that mammary gland homeostasis relies on unipotent stem cells. However, whether and when lineage restriction occurs during embryonic mammary development, and which signals orchestrate cell fate specification, remain unknown. Using a combination of in vivo clonal analysis with whole mount immunofluorescence and mathematical modelling of clonal dynamics, we found that embryonic multipotent mammary cells become lineage-restricted surprisingly early in development, with evidence for unipotency as early as E12.5 and no statistically discernable bipotency after E15.5. To gain insights into the mechanisms governing the switch from multipotency to unipotency, we used gain-of-function Notch1 mice and demonstrated that Notch activation cell autonomously dictates luminal cell fate specification to both embryonic and basally committed mammary cells. These functional studies have important implications for understanding the signals underlying cell plasticity and serve to clarify how reactivation of embryonic programs in adult cells can lead to cancer.
Conflict of interest statement
Figures

Comment in
-
Mammary lineage restriction in development.Nat Cell Biol. 2018 Jun;20(6):637-639. doi: 10.1038/s41556-018-0111-6. Nat Cell Biol. 2018. PMID: 29784916 No abstract available.
References
-
- Stingl J, Raouf A, Eirew P, Eaves CJ. Deciphering the mammary epithelial cell hierarchy. Cell Cycle. 2006;5:1519–1522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

