Early lineage segregation of multipotent embryonic mammary gland progenitors
- PMID: 29784918
- PMCID: PMC5985933
- DOI: 10.1038/s41556-018-0095-2
Early lineage segregation of multipotent embryonic mammary gland progenitors
Erratum in
-
Publisher Correction: Early lineage segregation of multipotent embryonic mammary gland progenitors.Nat Cell Biol. 2018 Sep;20(9):1099. doi: 10.1038/s41556-018-0164-6. Nat Cell Biol. 2018. PMID: 30018320
Abstract
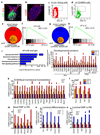
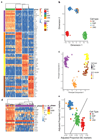
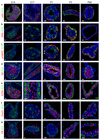
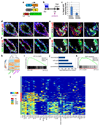
The mammary gland is composed of basal cells and luminal cells. It is generally believed that the mammary gland arises from embryonic multipotent progenitors, but it remains unclear when lineage restriction occurs and what mechanisms are responsible for the switch from multipotency to unipotency during its morphogenesis. Here, we perform multicolour lineage tracing and assess the fate of single progenitors, and demonstrate the existence of a developmental switch from multipotency to unipotency during embryonic mammary gland development. Molecular profiling and single cell RNA-seq revealed that embryonic multipotent progenitors express a unique hybrid basal and luminal signature and the factors associated with the different lineages. Sustained p63 expression in embryonic multipotent progenitors promotes unipotent basal cell fate and was sufficient to reprogram adult luminal cells into basal cells by promoting an intermediate hybrid multipotent-like state. Altogether, this study identifies the timing and the mechanisms mediating early lineage segregation of multipotent progenitors during mammary gland development.
Conflict of interest statement
The authors declare to have no financial and non-financial competing interests.
Figures



References
-
- Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. - PubMed
-
- de Visser KE, et al. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

