Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria
- PMID: 29784978
- PMCID: PMC5985962
- DOI: 10.1038/s41564-018-0162-2
Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria
Erratum in
-
Author Correction: Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria.Nat Microbiol. 2020 Oct;5(10):1306. doi: 10.1038/s41564-020-0787-9. Nat Microbiol. 2020. PMID: 32796922
Abstract
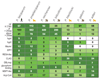
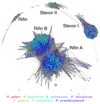
Plasmodium falciparum, the most virulent agent of human malaria, shares a recent common ancestor with the gorilla parasite Plasmodium praefalciparum. Little is known about the other gorilla- and chimpanzee-infecting species in the same (Laverania) subgenus as P. falciparum, but none of them are capable of establishing repeated infection and transmission in humans. To elucidate underlying mechanisms and the evolutionary history of this subgenus, we have generated multiple genomes from all known Laverania species. The completeness of our dataset allows us to conclude that interspecific gene transfers, as well as convergent evolution, were important in the evolution of these species. Striking copy number and structural variations were observed within gene families and one, stevor, shows a host-specific sequence pattern. The complete genome sequence of the closest ancestor of P. falciparum enables us to estimate the timing of the beginning of speciation to be 40,000-60,000 years ago followed by a population bottleneck around 4,000-6,000 years ago. Our data allow us also to search in detail for the features of P. falciparum that made it the only member of the Laverania able to infect and spread in humans.
Conflict of interest statement
None
Figures




Comment in
-
Evolution of human malaria.Nat Microbiol. 2018 Jun;3(6):642-643. doi: 10.1038/s41564-018-0170-2. Nat Microbiol. 2018. PMID: 29795536 No abstract available.
-
On the origin of Plasmodium falciparum.Nat Rev Microbiol. 2018 Jul;16(7):393. doi: 10.1038/s41579-018-0038-8. Nat Rev Microbiol. 2018. PMID: 29858596 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

