Genetic identification of brain cell types underlying schizophrenia
- PMID: 29785013
- PMCID: PMC6477180
- DOI: 10.1038/s41588-018-0129-5
Genetic identification of brain cell types underlying schizophrenia
Abstract
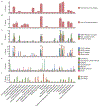
With few exceptions, the marked advances in knowledge about the genetic basis of schizophrenia have not converged on findings that can be confidently used for precise experimental modeling. By applying knowledge of the cellular taxonomy of the brain from single-cell RNA sequencing, we evaluated whether the genomic loci implicated in schizophrenia map onto specific brain cell types. We found that the common-variant genomic results consistently mapped to pyramidal cells, medium spiny neurons (MSNs) and certain interneurons, but far less consistently to embryonic, progenitor or glial cells. These enrichments were due to sets of genes that were specifically expressed in each of these cell types. We also found that many of the diverse gene sets previously associated with schizophrenia (genes involved in synaptic function, those encoding mRNAs that interact with FMRP, antipsychotic targets, etc.) generally implicated the same brain cell types. Our results suggest a parsimonious explanation: the common-variant genetic results for schizophrenia point at a limited set of neurons, and the gene sets point to the same cells. The genetic risk associated with MSNs did not overlap with that of glutamatergic pyramidal cells and interneurons, suggesting that different cell types have biologically distinct roles in schizophrenia.
Conflict of interest statement
Conflicts of Interest
The authors reports the following potentially competing financial interests. PF Sullivan: Lundbeck (advisory committee), Pfizer (Scientific Advisory Board member), and Roche (grant recipient, speaker reimbursement). J Hjerling-Leffler: Cartana (Scientific Adviser) and Roche (grant recipient).
Figures
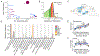

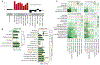

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

