Transcriptomic Impacts of Rumen Epithelium Induced by Butyrate Infusion in Dairy Cattle in Dry Period
- PMID: 29785087
- PMCID: PMC5954180
- DOI: 10.1177/1177625018774798
Transcriptomic Impacts of Rumen Epithelium Induced by Butyrate Infusion in Dairy Cattle in Dry Period
Abstract
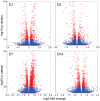
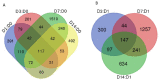
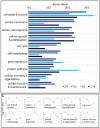
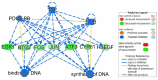
The purpose of this study was to evaluate the effects of butyrate infusion on rumen epithelial transcriptome. Next-generation sequencing (NGS) and bioinformatics are used to accelerate our understanding of regulation in rumen epithelial transcriptome of cattle in the dry period induced by butyrate infusion at the level of the whole transcriptome. Butyrate, as an essential element of nutrients, is a histone deacetylase (HDAC) inhibitor that can alter histone acetylation and methylation, and plays a prominent role in regulating genomic activities influencing rumen nutrition utilization and function. Ruminal infusion of butyrate was following 0-hour sampling (baseline controls) and continued for 168 hours at a rate of 5.0 L/day of a 2.5 M solution as a continuous infusion. Following the 168-hour infusion, the infusion was stopped, and cows were maintained on the basal lactation ration for an additional 168 hours for sampling. Rumen epithelial samples were serially collected via biopsy through rumen fistulae at 0-, 24-, 72-, and 168-hour (D1, D3, D7) and 168-hour post-infusion (D14). In comparison with pre-infusion at 0 hours, a total of 3513 genes were identified to be impacted in the rumen epithelium by butyrate infusion at least once at different sampling time points at a stringent cutoff of false discovery rate (FDR) < 0.01. The maximal effect of butyrate was observed at day 7. Among these impacted genes, 117 genes were responsive consistently from day 1 to day 14, and another 42 genes were lasting through day 7. Temporal effects induced by butyrate infusion indicate that the transcriptomic alterations are very dynamic. Gene ontology (GO) enrichment analysis revealed that in the early stage of rumen butyrate infusion (on day 1 and day 3 of butyrate infusion), the transcriptomic effects in the rumen epithelium were involved with mitotic cell cycle process, cell cycle process, and regulation of cell cycle. Bioinformatic analysis of cellular functions, canonical pathways, and upstream regulator of impacted genes underlie the potential mechanisms of butyrate-induced gene expression regulation in rumen epithelium. The introduction of transcriptomic and bioinformatic technologies to study nutrigenomics in the farm animal presented a new prospect to study multiple levels of biological information to better apprehend the whole animal response to nutrition, physiological state, and their interactions. The nutrigenomics approach may eventually lead to more precise management of utilization of feed resources in a more effective approach.
Keywords: Butyrate; Dairy Cattle; epithelium; rumen; transcriptome.
Conflict of interest statement
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

References
-
- Osorio JS, Vailati-Riboni M, Palladino A, Loor JJ. Application of nutrigenomics in small ruminants: lactation, growth, and beyond. Small Rumin Res. 2017;154:29–44.
-
- Goff JP, Horst RL. Physiological changes at parturition and their relationship to metabolic disorders. J Dairy Sci. 1997;80:1260–1268. - PubMed
-
- Duplessis M, Lapierre H, Ouattara B, et al. Whole-body propionate and glucose metabolism of multiparous dairy cows receiving folic acid and vitamin B12 supplements. J Dairy Sci. 2017;100:8578–8589. - PubMed
-
- Gálfi P, Neogrady S, Sakata T. Effects of volatile fatty acids on the epithelial cell proliferation of the digestive tract and its hormonal mediation. In: Tsuda T, ed. Proceedings of the Seventh International Symposium on Ruminant Physiology San Diego, CA: Academic Press; 1991:49–59.
-
- Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

