A study on the use of strain-specific and homologous promoters for heterologous expression in industrial Saccharomyces cerevisiae strains
- PMID: 29785587
- PMCID: PMC5962522
- DOI: 10.1186/s13568-018-0613-4
A study on the use of strain-specific and homologous promoters for heterologous expression in industrial Saccharomyces cerevisiae strains
Abstract
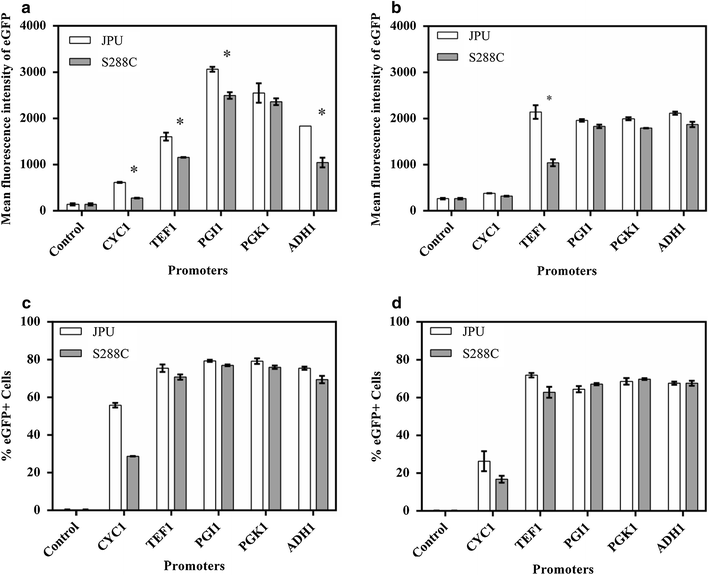
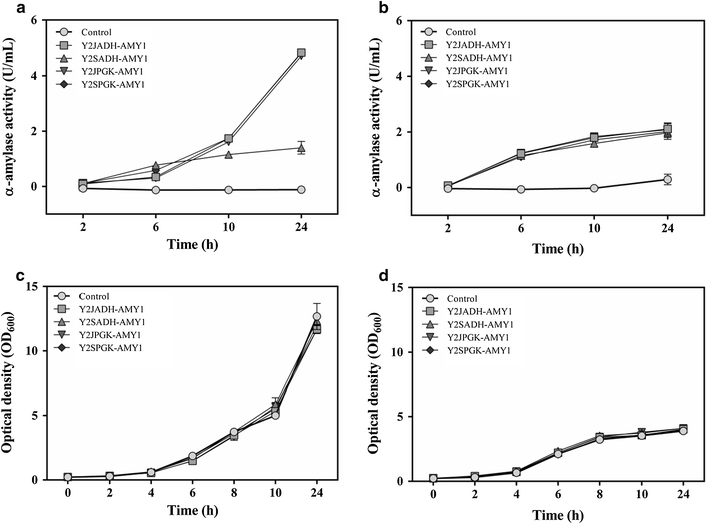
Polymorphism is well known in Saccharomyces cerevisiae strains used for different industrial applications, however little is known about its effects on promoter efficiency. In order to test this, five different promoters derived from an industrial and a laboratory (S288c) strain were used to drive the expression of eGFP reporter gene in both cells. The ADH1 promoter (P ADH1 ) in particular, which showed more polymorphism among the promoters analyzed, also exhibited the highest differences in intracellular fluorescence production. This was further confirmed by Northern blot analysis. The same behavior was also observed when the gene coding for secreted α-amylase from Cryptococcus flavus was placed under the control of either P ADH1 . These results underline the importance of the careful choice of the source of the promoter to be used in industrial yeast strains for heterologous expression.
Keywords: Amylase; Gene expression; Industrial yeast; Promoter; Saccharomyces cerevisiae.
Figures


References
-
- Akao T, Yashiro I, Hosoyama A, Kitagaki H, Horikawa H, Watanabe D, Akada R, Ando Y, Harashima S, Inoue T, Inoue Y, Kajiwara S, Kitamoto K, Kitamoto N, Kobayashi O, Kuhara S, Masubuchi T, Mizoguchi H, Nakao Y, Nakazato A, Namise M, Oba T, Ogata T, Ohta A, Sato M, Shibasaki S, Takatsume Y, Tanimoto S, Tsuboi H, Nishimura A, Yoda K, Ishikawa T, Iwashita K, Fujita N, Shimoil H. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 2011;18(6):423–434. doi: 10.1093/dnares/dsr029. - DOI - PMC - PubMed
-
- Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OVC, Missawa SK, Galzerani F, Costa GGL, Vidal RO, Noronha MF, Dominska M, Andrietta MGS, Andrietta SR, Cunha AF, Gomes LH, Tavares FCA, Alcarde AR, Dietrich FS, McCusker JH, Petes TD, Pereira GAG. Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res. 2009;19(12):2258–2270. doi: 10.1101/gr.091777.109. - DOI - PMC - PubMed
-
- Babrzadeh F, Jalili R, Wang C, Shokralla S, Pierce S, Robinson-Mosher A, Nyren P, Shafer RW, Basso LC, de Amorim HV, de Oliveira AJ, Davis RW, Ronaghi M, Gharizadeh B, Stambuk BU. Whole-genome sequencing of the efficient industrial fuel-ethanol fermentative Saccharomyces cerevisiae strain CAT-1. Mol Genet Genomics. 2012;287(6):485–494. doi: 10.1007/s00438-012-0695-7. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

