Comparison of Two Genotyping Methods for Distinguishing Recrudescence from Reinfection in Antimalarial Drug Efficacy/Effectiveness Trials
- PMID: 29785925
- PMCID: PMC6085787
- DOI: 10.4269/ajtmh.18-0002
Comparison of Two Genotyping Methods for Distinguishing Recrudescence from Reinfection in Antimalarial Drug Efficacy/Effectiveness Trials
Abstract
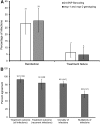
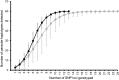
Genotyping of allelic variants of Plasmodium falciparum merozoite surface proteins 1 and 2 (msp-1 and msp-2), and the glutamate-rich protein is the gold standard for distinguishing reinfections from recrudescences in antimalarial drug trials. We compared performance of the recently developed 24-single-nucleotide polymorphism (SNP) Barcoding Assay against msp-1 and msp-2 genotyping in a cluster-randomized effectiveness trial of artemether-lumefantrine and dihydroartemisinin-piperaquine in Malawi. Rates of recrudescence and reinfection estimated by the two methods did not differ significantly (Fisher's exact test; P = 0.887 and P = 0.768, respectively). There was a strong agreement between the two methods in predicting treatment outcomes and resolving the genetic complexity of malaria infections in this setting. These results support the use of this SNP assay as an alternative method for correcting antimalarial efficacy/effectiveness data.
Figures
References
-
- World Health Organization , 2008. Methods and Techniques for Clinical Trials on Antimalarial Drug Efficacy: Fenotyping to Identify Parasite Populations. Informal Consultation Organized by the Medicines for Malaria Venture and Cosponsored by the World Health Organization. May 29–31, 2007, Amsterdam, The Netherlands.
-
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S, 1999. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg 93: 369–374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

