The draft genome sequence of cork oak
- PMID: 29786699
- PMCID: PMC5963338
- DOI: 10.1038/sdata.2018.69
The draft genome sequence of cork oak
Abstract
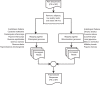
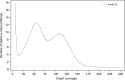
Cork oak (Quercus suber) is native to southwest Europe and northwest Africa where it plays a crucial environmental and economical role. To tackle the cork oak production and industrial challenges, advanced research is imperative but dependent on the availability of a sequenced genome. To address this, we produced the first draft version of the cork oak genome. We followed a de novo assembly strategy based on high-throughput sequence data, which generated a draft genome comprising 23,347 scaffolds and 953.3 Mb in size. A total of 79,752 genes and 83,814 transcripts were predicted, including 33,658 high-confidence genes. An InterPro signature assignment was detected for 69,218 transcripts, which represented 82.6% of the total. Validation studies demonstrated the genome assembly and annotation completeness and highlighted the usefulness of the draft genome for read mapping of high-throughput sequence data generated using different protocols. All data generated is available through the public databases where it was deposited, being therefore ready to use by the academic and industry communities working on cork oak and/or related species.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Data Citations
-
- 2018. GenBank. PKMF00000000
-
- 2017. NCBI Sequence Read Archive. SRP111728
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

