A Bayesian hidden Potts mixture model for analyzing lung cancer pathology images
- PMID: 29788035
- PMCID: PMC6797059
- DOI: 10.1093/biostatistics/kxy019
A Bayesian hidden Potts mixture model for analyzing lung cancer pathology images
Abstract
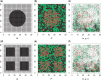
Digital pathology imaging of tumor tissues, which captures histological details in high resolution, is fast becoming a routine clinical procedure. Recent developments in deep-learning methods have enabled the identification, characterization, and classification of individual cells from pathology images analysis at a large scale. This creates new opportunities to study the spatial patterns of and interactions among different types of cells. Reliable statistical approaches to modeling such spatial patterns and interactions can provide insight into tumor progression and shed light on the biological mechanisms of cancer. In this article, we consider the problem of modeling a pathology image with irregular locations of three different types of cells: lymphocyte, stromal, and tumor cells. We propose a novel Bayesian hierarchical model, which incorporates a hidden Potts model to project the irregularly distributed cells to a square lattice and a Markov random field prior model to identify regions in a heterogeneous pathology image. The model allows us to quantify the interactions between different types of cells, some of which are clinically meaningful. We use Markov chain Monte Carlo sampling techniques, combined with a double Metropolis-Hastings algorithm, in order to simulate samples approximately from a distribution with an intractable normalizing constant. The proposed model was applied to the pathology images of $205$ lung cancer patients from the National Lung Screening trial, and the results show that the interaction strength between tumor and stromal cells predicts patient prognosis (P = $0.005$). This statistical methodology provides a new perspective for understanding the role of cell-cell interactions in cancer progression.
Keywords: Double Metropolis–Hastings; Hidden Potts model; Lung cancer; Markov random field; Mixture model; Pathology image; Potts model; Spatial point pattern.
© The Author 2018. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Amin, M. B., Tamboli, P., Merchant, S. H., Ordóñez, N. G., Ro, J., Ayala, A. G. and Ro, J. Y. (2002). Micropapillary component in lung adenocarcinoma: a distinctive histologic feature with possible prognostic significance. The American Journal of Surgical Pathology 26, 358–364. - PubMed
-
- Ayasso, H. and Mohammad-Djafari, A. (2010). Joint NDT image restoration and segmentation using Gauss–Markov–Potts prior models and variational Bayesian computation. IEEE Transactions on Image Processing 19, 2265–2277. - PubMed
-
- Beck, A. H., Sangoi, A. R., Leung, S., Marinelli, R. J., Nielsen, T. O., van de Vijver, M. J., West, R. B., van de Rijn, M. and Koller, D. (2011). Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Science Translational Medicine 3, 108ra113. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

