Constitutive Interferon Pathway Activation in Tumors as an Efficacy Determinant Following Oncolytic Virotherapy
- PMID: 29788332
- PMCID: PMC6186520
- DOI: 10.1093/jnci/djy033
Constitutive Interferon Pathway Activation in Tumors as an Efficacy Determinant Following Oncolytic Virotherapy
Abstract
Background: Attenuated measles virus (MV) strains are promising agents currently being tested against solid tumors or hematologic malignancies in ongoing phase I and II clinical trials; factors determining oncolytic virotherapy success remain poorly understood, however.
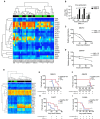
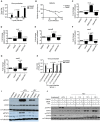
Methods: We performed RNA sequencing and gene set enrichment analysis to identify pathways differentially activated in MV-resistant (n = 3) and -permissive (n = 2) tumors derived from resected human glioblastoma (GBM) specimens and propagated as xenografts (PDX). Using a unique gene signature we identified, we generated a diagonal linear discriminant analysis (DLDA) classification algorithm to predict MV responders and nonresponders, which was validated in additional randomly selected GBM and ovarian cancer PDX and 10 GBM patients treated with MV in a phase I trial. GBM PDX lines were also treated with the US Food and Drug Administration-approved JAK inhibitor, ruxolitinib, for 48 hours prior to MV infection and virus production, STAT1/3 signaling and interferon stimulated gene expression was assessed. All statistical tests were two-sided.
Results: Constitutive interferon pathway activation, as reflected in the DLDA algorithm, was identified as the key determinant for MV replication, independent of virus receptor expression, in MV-permissive and -resistant GBM PDXs. Using these lines as the training data for the DLDA algorithm, we confirmed the accuracy of our algorithm in predicting MV response in randomly selected GBM PDX ovarian cancer PDXs. Using the DLDA prediction algorithm, we demonstrate that virus replication in patient tumors is inversely correlated with expression of this resistance gene signature (ρ = -0.717, P = .03). In vitro inhibition of the interferon response pathway with the JAK inhibitor ruxolitinib was able to overcome resistance and increase virus production (1000-fold, P = .03) in GBM PDX lines.
Conclusions: These findings document a key mechanism of tumor resistance to oncolytic MV therapy and describe for the first time the development of a prediction algorithm to preselect for oncolytic treatment or combinatorial strategies.
Figures



Comment in
-
Viral Therapy Gets Personal: A Potential Gene Signature to Predict Susceptibility to Measles Virus Oncolysis.J Natl Cancer Inst. 2018 Oct 1;110(10):1139-1140. doi: 10.1093/jnci/djy061. J Natl Cancer Inst. 2018. PMID: 29757401 No abstract available.
References
-
- Andtbacka RH, Kaufman HL, Collichio F, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;3325:2780–2788. - PubMed
-
- Dorig RE, Marcil A, Chopra A, et al. The human Cd46 molecule is a receptor for measles-virus (Edmonston strain). Cell. 1993;752:295–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

