Hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat
- PMID: 29788353
- PMCID: PMC6054230
- DOI: 10.1093/jxb/ery190
Hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat
Abstract
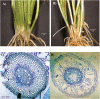
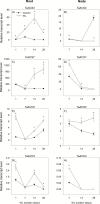
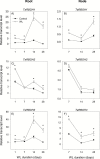
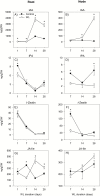
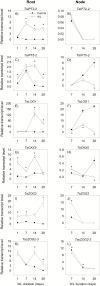
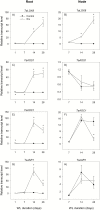
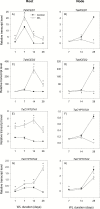
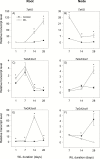
To gain insights into the molecular mechanisms underlying hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat, the present study investigated transcriptional regulation of genes related to hormone metabolism and transport in the root and stem node tissues. Waterlogging-induced inhibition of axile root elongation and lateral root formation, and promotion of surface adventitious and axile root emergence and aerenchyma formation are associated with enhanced expression levels of ethylene biosynthesis genes, ACS7 and ACO2, in both tissues. Inhibition of axile root elongation is also related to increased root indole acetic acid (IAA) and jasmonate (JA) levels that are associated with up-regulation of specific IAA biosynthesis/transport (TDC, YUC1, and PIN9) and JA metabolism (LOX8, AOS1, AOC1, and JAR1) genes, and transcriptional alteration of gibberellin (GA) metabolism genes (GA3ox2 and GA2ox8). Adventitious root emergence from waterlogged stem nodes is associated with increased levels of IAA and GA but decreased levels of cytokinin and abscisic acid (ABA), which are regulated through the expression of specific IAA biosynthesis/transport (TDC, YUC1, and PIN9), cytokinin metabolism (IPT5-2, LOG1, CKX5, and ZOG2), ABA biosynthesis (NCED1 and NCED2), and GA metabolism (GA3ox2 and GA2ox8) genes. These results enhance our understanding of the molecular mechanisms underlying the adaptive response of wheat to waterlogging.
Figures



References
-
- Arbona V, Gómez-Cadenas A. 2008. Hormonal modulation of citrus responses to flooding. Journal of Plant Growth Regulation 27, 241.
-
- Armstrong W. 1979. Aeration in higher plants. In: Woolhouse HW, ed. Advances in botanical research. London: Academic Press, 225–332.
-
- Blilou I, Xu J, Wildwater M, et al. . 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

