Antisense transcriptional interference mediates condition-specific gene repression in budding yeast
- PMID: 29788449
- PMCID: PMC6158615
- DOI: 10.1093/nar/gky342
Antisense transcriptional interference mediates condition-specific gene repression in budding yeast
Abstract
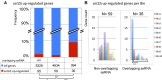
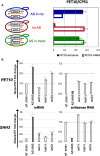
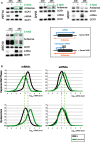
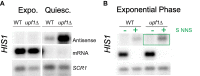
Pervasive transcription generates many unstable non-coding transcripts in budding yeast. The transcription of such noncoding RNAs, in particular antisense RNAs (asRNAs), has been shown in a few examples to repress the expression of the associated mRNAs. Yet, such mechanism is not known to commonly contribute to the regulation of a given class of genes. Using a mutant context that stabilized pervasive transcripts, we observed that the least expressed mRNAs during the exponential phase were associated with high levels of asRNAs. These asRNAs also overlapped their corresponding gene promoters with a much higher frequency than average. Interrupting antisense transcription of a subset of genes corresponding to quiescence-enriched mRNAs restored their expression. The underlying mechanism acts in cis and involves several chromatin modifiers. Our results convey that transcription interference represses up to 30% of the 590 least expressed genes, which includes 163 genes with quiescence-enriched mRNAs. We also found that pervasive transcripts constitute a higher fraction of the transcriptome in quiescence relative to the exponential phase, consistent with gene expression itself playing an important role to suppress pervasive transcription. Accordingly, the HIS1 asRNA, normally only present in quiescence, is expressed in exponential phase upon HIS1 mRNA transcription interruption.
Figures




References
-
- Carninci P., Hayashizaki Y.. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 2007; 17:139–144. - PubMed
-
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M. et al. . Life with 6000 genes. Science. 1996; 274:563–567. - PubMed
-
- Jensen T.H., Jacquier A., Libri D.. Dealing with pervasive transcription. Mol. Cell. 2013; 52:473–484. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

