CellAtlasSearch: a scalable search engine for single cells
- PMID: 29788498
- PMCID: PMC6030823
- DOI: 10.1093/nar/gky421
CellAtlasSearch: a scalable search engine for single cells
Abstract
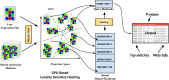
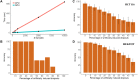
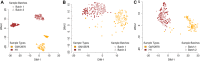
Owing to the advent of high throughput single cell transcriptomics, past few years have seen exponential growth in production of gene expression data. Recently efforts have been made by various research groups to homogenize and store single cell expression from a large number of studies. The true value of this ever increasing data deluge can be unlocked by making it searchable. To this end, we propose CellAtlasSearch, a novel search architecture for high dimensional expression data, which is massively parallel as well as light-weight, thus infinitely scalable. In CellAtlasSearch, we use a Graphical Processing Unit (GPU) friendly version of Locality Sensitive Hashing (LSH) for unmatched speedup in data processing and query. Currently, CellAtlasSearch features over 300 000 reference expression profiles including both bulk and single-cell data. It enables the user query individual single cell transcriptomes and finds matching samples from the database along with necessary meta information. CellAtlasSearch aims to assist researchers and clinicians in characterizing unannotated single cells. It also facilitates noise free, low dimensional representation of single-cell expression profiles by projecting them on a wide variety of reference samples. The web-server is accessible at: http://www.cellatlassearch.com.
Figures

References
-
- Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Oudenaarden A.. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015; 525:251. - PubMed
-
- Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P. et al. . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010; 17:98–110. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

