Genetic diversity of BoLA-DRB3 in South American Zebu cattle populations
- PMID: 29788904
- PMCID: PMC5964877
- DOI: 10.1186/s12863-018-0618-7
Genetic diversity of BoLA-DRB3 in South American Zebu cattle populations
Abstract
Background: Bovine leukocyte antigens (BoLAs) are used extensively as markers of disease and immunological traits in cattle. However, until now, characterization of BoLA gene polymorphisms in Zebu breeds using high resolution typing methods has been poor. Here, we used a polymerase chain reaction sequence-based typing (PCR-SBT) method to sequence exon 2 of the BoLA class II DRB3 gene from 421 cattle (116 Bolivian Nellore, 110 Bolivian Gir, and 195 Peruvian Nellore-Brahman). Data from 1416 Taurine and Zebu samples were also included in the analysis.
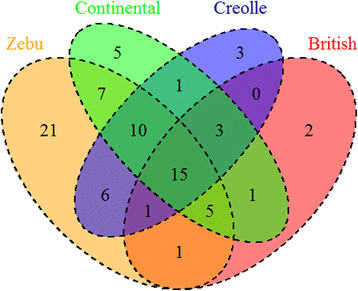
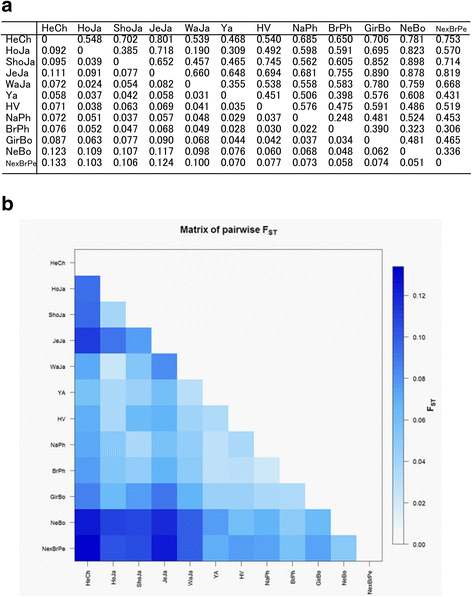
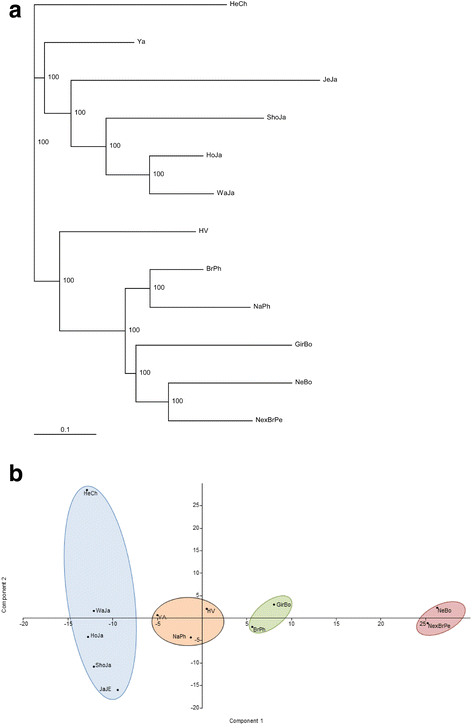
Results: We identified 46 previously reported alleles and no novel variants. Of note, 1/3 of the alleles were detected only in Zebu cattle. Comparison of the degree of genetic variability at the population and sequence levels with genetic distance in the three above mentioned breeds and nine previously reported breeds revealed that Zebu breeds had a gene diversity score higher than 0.86, a nucleotide diversity score higher than 0.06, and a mean number of pairwise differences greater than 16, being similar to those estimated for other cattle breeds. A neutrality test revealed that only Nellore-Brahman cattle showed the even gene frequency distribution expected under a balanced selection scenario. The FST index and the exact G test showed significant differences across all cattle populations (FST = 0.057; p < 0.001). Neighbor-joining trees and principal component analysis identified two major clusters: one comprising mainly European Taurine breeds and a second comprising Zebu breeds. This is consistent with the historical and geographical origin of these breeds. Some of these differences may be explained by variation of amino acid motifs at antigen-binding sites.
Conclusions: The results presented herein show that the historical divergence between Taurine and Zebu cattle breeds is a result of origin, selection, and adaptation events, which would explain the observed differences in BoLA-DRB3 gene diversity between the two major bovine types. This allelic information will be important for investigating the relationship between the major histocompatibility complex and disease, and contribute to an ongoing effort to catalog bovine MHC allele frequencies according to breed and location.
Keywords: BoLA-DRB3; Brahman; Genetic diversity; Gir; Nellore; Sequence-based typing.
Conflict of interest statement
Ethics approval
All animals were handled by veterinarians from RIKEN, Universidad Austral de Chile and LAVET, in strict accordance with good animal practice following the Universidad Austral de Chile Institutional guidelines. This study was approved by the Committee on the Ethics of Animals for Research at the National University of LA PLATA (Certificate date May 26th, 2014) and by the Committee on the Ethics of Animals for Research at Universidad Austral de Chile (Certificate No. 153–2014). The animal has derived from private owners and the consent was taken in verbal form, because each farmer commonly contacts with veterinarian in same way.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Characterization of bovine MHC DRB3 diversity in global cattle breeds, with a focus on cattle in Myanmar.BMC Genet. 2020 Sep 1;21(1):95. doi: 10.1186/s12863-020-00905-8. BMC Genet. 2020. PMID: 32867670 Free PMC article.
-
Characterization of bovine MHC DRB3 diversity in Latin American Creole cattle breeds.Gene. 2013 Apr 25;519(1):150-8. doi: 10.1016/j.gene.2013.01.002. Epub 2013 Jan 16. Gene. 2013. PMID: 23333729
-
Characterization of bovine MHC class II DRB3 diversity in South American Holstein cattle populations.Tissue Antigens. 2015 Dec;86(6):419-30. doi: 10.1111/tan.12692. Epub 2015 Oct 30. Tissue Antigens. 2015. PMID: 26514650
-
Graduate Student Literature Review: The DRB3 gene of the bovine major histocompatibility complex-Discovery, diversity, and distribution of alleles in commercial breeds of cattle and applications for development of vaccines.J Dairy Sci. 2024 Dec;107(12):11324-11341. doi: 10.3168/jds.2023-24628. Epub 2024 Jul 14. J Dairy Sci. 2024. PMID: 39004123 Review.
-
Evolution and domestication of the Bovini species.Anim Genet. 2020 Oct;51(5):637-657. doi: 10.1111/age.12974. Epub 2020 Jul 27. Anim Genet. 2020. PMID: 32716565 Review.
Cited by
-
Nanovaccines against Animal Pathogens: The Latest Findings.Vaccines (Basel). 2021 Sep 4;9(9):988. doi: 10.3390/vaccines9090988. Vaccines (Basel). 2021. PMID: 34579225 Free PMC article. Review.
-
Genetic Variation and Population Differentiation in the Bovine Lymphocyte Antigen DRB3.2 Locus of South African Nguni Crossbred Cattle.Animals (Basel). 2021 Jun 2;11(6):1651. doi: 10.3390/ani11061651. Animals (Basel). 2021. PMID: 34199370 Free PMC article.
-
Molecular Characteristics and Processing Technologies of Dairy Products from Non-Traditional Species.Molecules. 2024 Nov 18;29(22):5427. doi: 10.3390/molecules29225427. Molecules. 2024. PMID: 39598816 Free PMC article. Review.
-
Genetic Diversity and Sequence Conservation of Peptide-Binding Regions of MHC Class I Genes in Pig, Cattle, Chimpanzee, and Human.Genes (Basel). 2023 Dec 20;15(1):7. doi: 10.3390/genes15010007. Genes (Basel). 2023. PMID: 38275589 Free PMC article.
-
A pooled testing system to rapidly identify cattle carrying the elite controller BoLA-DRB3*009:02 haplotype against bovine leukemia virus infection.HLA. 2022 Jan;99(1):12-24. doi: 10.1111/tan.14502. Epub 2021 Dec 19. HLA. 2022. PMID: 34837483 Free PMC article.
References
-
- Aida Y. Characterization and expression of bovine MHC class II genes. Bull Soc Fr Jpn Sci Vet. 1995;6:17–24.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

