Whole genome analysis reveals the diversity and evolutionary relationships between necrotic enteritis-causing strains of Clostridium perfringens
- PMID: 29788909
- PMCID: PMC5964661
- DOI: 10.1186/s12864-018-4771-1
Whole genome analysis reveals the diversity and evolutionary relationships between necrotic enteritis-causing strains of Clostridium perfringens
Abstract
Background: Clostridium perfringens causes a range of diseases in animals and humans including necrotic enteritis in chickens and food poisoning and gas gangrene in humans. Necrotic enteritis is of concern in commercial chicken production due to the cost of the implementation of infection control measures and to productivity losses. This study has focused on the genomic analysis of a range of chicken-derived C. perfringens isolates, from around the world and from different years. The genomes were sequenced and compared with 20 genomes available from public databases, which were from a diverse collection of isolates from chickens, other animals, and humans. We used a distance based phylogeny that was constructed based on gene content rather than sequence identity. Similarity between strains was defined as the number of genes that they have in common divided by their total number of genes. In this type of phylogenetic analysis, evolutionary distance can be interpreted in terms of evolutionary events such as acquisition and loss of genes, whereas the underlying properties (the gene content) can be interpreted in terms of function. We also compared these methods to the sequence-based phylogeny of the core genome.
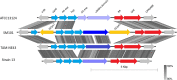
Results: Distinct pathogenic clades of necrotic enteritis-causing C. perfringens were identified. They were characterised by variable regions encoded on the chromosome, with predicted roles in capsule production, adhesion, inhibition of related strains, phage integration, and metabolism. Some strains have almost identical genomes, even though they were isolated from different geographic regions at various times, while other highly distant genomes appear to result in similar outcomes with regard to virulence and pathogenesis.
Conclusions: The high level of diversity in chicken isolates suggests there is no reliable factor that defines a chicken strain of C. perfringens, however, disease-causing strains can be defined by the presence of netB-encoding plasmids. This study reveals that horizontal gene transfer appears to play a significant role in genetic variation of the C. perfringens chromosome as well as the plasmid content within strains.
Keywords: Adhesion; Capsule; Clostridium perfringens; Genome; Necrotic enteritis; Pangenome; Prophage.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable. All the work reported is laboratory based, not requiring human or animal ethics approvals or consents.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures









Similar articles
-
Genomic diversity of necrotic enteritis-associated strains of Clostridium perfringens: a review.Avian Pathol. 2016 Jun;45(3):302-7. doi: 10.1080/03079457.2016.1153799. Avian Pathol. 2016. PMID: 26949841 Review.
-
Conjugation-Mediated Horizontal Gene Transfer of Clostridium perfringens Plasmids in the Chicken Gastrointestinal Tract Results in the Formation of New Virulent Strains.Appl Environ Microbiol. 2017 Dec 1;83(24):e01814-17. doi: 10.1128/AEM.01814-17. Print 2017 Dec 15. Appl Environ Microbiol. 2017. PMID: 29030439 Free PMC article.
-
Sialidase production and genetic diversity in Clostridium perfringens type A isolated from chicken with necrotic enteritis in Brazil.Curr Microbiol. 2015 Mar;70(3):330-7. doi: 10.1007/s00284-014-0722-5. Epub 2014 Nov 6. Curr Microbiol. 2015. PMID: 25373329
-
Necrotic enteritis in broilers: an updated review on the pathogenesis.Avian Pathol. 2011 Aug;40(4):341-7. doi: 10.1080/03079457.2011.590967. Avian Pathol. 2011. PMID: 21812711 Review.
-
Plasmid Characterization and Chromosome Analysis of Two netF+ Clostridium perfringens Isolates Associated with Foal and Canine Necrotizing Enteritis.PLoS One. 2016 Feb 9;11(2):e0148344. doi: 10.1371/journal.pone.0148344. eCollection 2016. PLoS One. 2016. PMID: 26859667 Free PMC article.
Cited by
-
Focal duodenal necrosis in chickens: attempts to reproduce the disease experimentally and diagnostic considerations.J Vet Diagn Invest. 2020 Mar;32(2):268-276. doi: 10.1177/1040638720901726. Epub 2020 Jan 26. J Vet Diagn Invest. 2020. PMID: 31983302 Free PMC article.
-
Pathogenicity and virulence of Clostridium perfringens.Virulence. 2021 Dec;12(1):723-753. doi: 10.1080/21505594.2021.1886777. Virulence. 2021. PMID: 33843463 Free PMC article.
-
Single and multiplexed gene repression in solventogenic Clostridium via Cas12a-based CRISPR interference.Synth Syst Biotechnol. 2022 Dec 24;8(1):148-156. doi: 10.1016/j.synbio.2022.12.005. eCollection 2023 Mar. Synth Syst Biotechnol. 2022. PMID: 36687471 Free PMC article.
-
Putative antigenic proteins identified by comparative and subtractive reverse vaccinology in necrotic enteritis-causing Clostridium perfringens isolated from broiler chickens.BMC Genomics. 2021 Dec 13;22(1):890. doi: 10.1186/s12864-021-08216-7. BMC Genomics. 2021. PMID: 34903179 Free PMC article.
-
Large-Scale Genomic Analyses and Toxinotyping of Clostridium perfringens Implicated in Foodborne Outbreaks in France.Front Microbiol. 2019 Apr 17;10:777. doi: 10.3389/fmicb.2019.00777. eCollection 2019. Front Microbiol. 2019. PMID: 31057505 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

