Concepts and Methods to Access Novel Antibiotics from Actinomycetes
- PMID: 29789481
- PMCID: PMC6022970
- DOI: 10.3390/antibiotics7020044
Concepts and Methods to Access Novel Antibiotics from Actinomycetes
Abstract
Actinomycetes have been proven to be an excellent source of secondary metabolites for more than half a century. Exhibiting various bioactivities, they provide valuable approved drugs in clinical use. Most microorganisms are still untapped in terms of their capacity to produce secondary metabolites, since only a small fraction can be cultured in the laboratory. Thus, improving cultivation techniques to extend the range of secondary metabolite producers accessible under laboratory conditions is an important first step in prospecting underexplored sources for the isolation of novel antibiotics. Currently uncultured actinobacteria can be made available by bioprospecting extreme or simply habitats other than soil. Furthermore, bioinformatic analysis of genomes reveals most producers to harbour many more biosynthetic gene clusters than compounds identified from any single strain, which translates into a silent biosynthetic potential of the microbial world for the production of yet unknown natural products. This review covers discovery strategies and innovative methods recently employed to access the untapped reservoir of natural products. The focus is the order of actinomycetes although most approaches are similarly applicable to other microbes. Advanced cultivation methods, genomics- and metagenomics-based approaches, as well as modern metabolomics-inspired methods are highlighted to emphasise the interplay of different disciplines to improve access to novel natural products.
Keywords: dereplication; genome mining; metabolomics; metagenomics; natural products; rare actinomycetes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


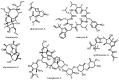


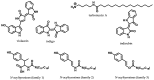

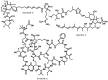
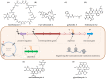

References
-
- World Health Organization Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation. [(accessed on 31 January 2018)]; Available online: http://apps.who.int/iris/bitstream/handle/10665/259744/9789241513449-eng....
-
- Luzhetskyy A., Pelzer S., Bechthold A. The future of natural products as a source of new antibiotics. Curr. Opin. Investig. Drugs. 2007;8:608–613. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

