Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods
- PMID: 29789543
- PMCID: PMC5964202
- DOI: 10.1038/s41467-018-04254-0
Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods
Abstract
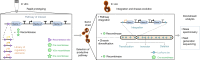
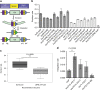
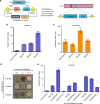
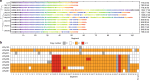
Exogenous pathway optimization and chassis engineering are two crucial methods for heterologous pathway expression. The two methods are normally carried out step-wise and in a trial-and-error manner. Here we report a recombinase-based combinatorial method (termed "SCRaMbLE-in") to tackle both challenges simultaneously. SCRaMbLE-in includes an in vitro recombinase toolkit to rapidly prototype and diversify gene expression at the pathway level and an in vivo genome reshuffling system to integrate assembled pathways into the synthetic yeast genome while combinatorially causing massive genome rearrangements in the host chassis. A set of loxP mutant pairs was identified to maximize the efficiency of the in vitro diversification. Exemplar pathways of β-carotene and violacein were successfully assembled, diversified, and integrated using this SCRaMbLE-in method. High-throughput sequencing was performed on selected engineered strains to reveal the resulting genotype-to-phenotype relationships. The SCRaMbLE-in method proves to be a rapid, efficient, and universal method to fast track the cycle of engineering biology.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

