Long non-coding RNA Gm2199 rescues liver injury and promotes hepatocyte proliferation through the upregulation of ERK1/2
- PMID: 29789577
- PMCID: PMC5964236
- DOI: 10.1038/s41419-018-0595-9
Long non-coding RNA Gm2199 rescues liver injury and promotes hepatocyte proliferation through the upregulation of ERK1/2
Abstract
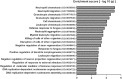
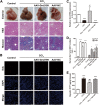
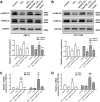
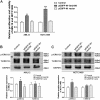
Long non-coding RNAs (lncRNAs) are a new class of regulators of various human diseases. This study was designed to explore the potential role of lncRNAs in experimental hepatic damage. In vivo hepatic damage in mice and in vitro hepatocyte damage in AML12 and NCTC1469 cells were induced by carbon tetrachloride (CCl4) treatments. Expression profiles of lncRNAs and mRNAs were analyzed by microarray. Bioinformatics analyses were conducted to predict the potential functions of differentially expressed lncRNAs with respect to hepatic damage. Overexpression of lncRNA Gm2199 was achieved by transfection of the pEGFP-N1-Gm2199 plasmid in vitro and adeno-associated virus-Gm2199 in vivo. Cell proliferation and viability was detected by cell counting kit-8 and 5-ethynyl-2'-deoxyuridine assay. Protein and mRNA expressions of extracellular signal-regulated kinase-1/2 (ERK1/2) were detected by western blot and quantitative real-time reverse-transcription PCR (qRT-PCR). Microarray analysis identified 190 and 148 significantly differentially expressed lncRNAs and mRNAs, respectively. The analyses of lncRNA-mRNA co-expression and lncRNA-biological process networks unraveled potential roles of the differentially expressed lncRNAs including Gm2199 in the pathophysiological processes leading to hepatic damage. Gm2199 was downregulated in both damaged livers and hepatocyte lines. Overexpression of Gm2199 restored the reduced proliferation of damaged hepatocyte lines and increased the expression of ERK1/2. Overexpression of Gm2199 also promoted the proliferation and viability of normal hepatocyte lines and increased the level of p-ERK1/2. Overexpression of Gm2199 in vivo also protected mouse liver injury induced by CCl4, evidenced by more proliferating hepatocytes, less serum alanine aminotransferase, less serum aspartate aminotransferase, and decreased hepatic hydroxyproline. The ability of Gm2199 to maintain hepatic proliferation capacity indicates it as a novel anti-liver damage lncRNA.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

