ClinVar Miner: Demonstrating utility of a Web-based tool for viewing and filtering ClinVar data
- PMID: 29790234
- PMCID: PMC6043391
- DOI: 10.1002/humu.23555
ClinVar Miner: Demonstrating utility of a Web-based tool for viewing and filtering ClinVar data
Abstract
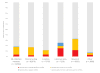
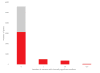
ClinVar Miner is a Web-based suite that utilizes the data held in the National Center for Biotechnology Information's ClinVar archive. The goal is to render the data more accessible to processes pertaining to conflict resolution of variant interpretation as well as tracking details of data submission and data management for detailed variant curation. Here, we establish the use of these tools to address three separate use cases and to perform analyses across submissions. We demonstrate that the ClinVar Miner tools are an effective means to browse and consolidate data for variant submitters, curation groups, and general oversight. These tools are also relevant to the variant interpretation community in general.
Keywords: ClinVar; clinical domain working group; expert panel; variant archive; variant curation; variant interpretation.
© 2018 Wiley Periodicals, Inc.
Figures





References
-
- Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mefford HC, … Ledbetter N, editors. GeneReviews(®) Seattle (WA): University of Washington, Seattle; 2010.
-
- Clinical Laboratories Meeting Minimum Requirements for Data Sharing to Support Quality Assurance - ClinGen. Clinical Genome Resource. n.d Retrieved February 6, 2018, from https://www.clinicalgenome.org/lablist/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

