PyCoTools: a Python toolbox for COPASI
- PMID: 29790940
- PMCID: PMC6198863
- DOI: 10.1093/bioinformatics/bty409
PyCoTools: a Python toolbox for COPASI
Abstract
Motivation: COPASI is an open source software package for constructing, simulating and analyzing dynamic models of biochemical networks. COPASI is primarily intended to be used with a graphical user interface but often it is desirable to be able to access COPASI features programmatically, with a high level interface.
Results: PyCoTools is a Python package aimed at providing a high level interface to COPASI tasks with an emphasis on model calibration. PyCoTools enables the construction of COPASI models and the execution of a subset of COPASI tasks including time courses, parameter scans and parameter estimations. Additional 'composite' tasks which use COPASI tasks as building blocks are available for increasing parameter estimation throughput, performing identifiability analysis and performing model selection. PyCoTools supports exploratory data analysis on parameter estimation data to assist with troubleshooting model calibrations. We demonstrate PyCoTools by posing a model selection problem designed to show case PyCoTools within a realistic scenario. The aim of the model selection problem is to test the feasibility of three alternative hypotheses in explaining experimental data derived from neonatal dermal fibroblasts in response to TGF-β over time. PyCoTools is used to critically analyze the parameter estimations and propose strategies for model improvement.
Availability and implementation: PyCoTools can be downloaded from the Python Package Index (PyPI) using the command 'pip install pycotools' or directly from GitHub (https://github.com/CiaranWelsh/pycotools). Documentation at http://pycotools.readthedocs.io.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

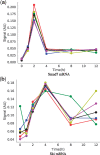
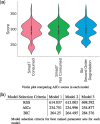
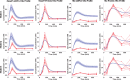
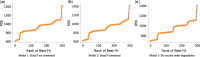
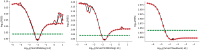
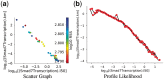
References
-
- Akiyoshi S. et al. (1999) c-ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with smads. J. Biol. Chem., 274, 35269–35277. - PubMed
-
- Choi K. et al. (2016) Tellurium: a python based modeling and reproducibility platform for systems biology. bioRxiv, 054601.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

