Identifying and avoiding off-target effects of RNase H-dependent antisense oligonucleotides in mice
- PMID: 29790953
- PMCID: PMC6009603
- DOI: 10.1093/nar/gky397
Identifying and avoiding off-target effects of RNase H-dependent antisense oligonucleotides in mice
Abstract
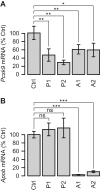
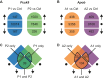
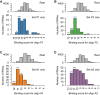
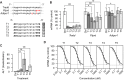
Antisense oligonucleotides that are dependent on RNase H for cleavage and subsequent degradation of complementary RNA are being developed as therapeutics. Besides the intended RNA target, such oligonucleotides may also cause degradation of unintended RNA off-targets by binding to partially complementary target sites. Here, we characterized the global effects on the mouse liver transcriptome of four oligonucleotides designed as gapmers, two targeting Apob and two targeting Pcsk9, all in different regions on their respective intended targets. This study design allowed separation of intended- and off-target effects on the transcriptome for each gapmer. Next, we used sequence analysis to identify possible partially complementary binding sites among the potential off-targets, and validated these by measurements of melting temperature and RNase H-cleavage rates. Generally, our observations were as expected in that fewer mismatches or bulges in the gapmer/transcript duplexes resulted in a higher chance of those duplexes being effective substrates for RNase H. Follow-up experiments in mice and cells show, that off-target effects can be mitigated by ensuring that gapmers have minimal sequence complementarity to any RNA besides the intended target, and that they do not have exaggerated binding affinity to the intended target.
Figures




References
-
- Stein H., Hausen P.. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: Effect on DNA-Dependent RNA polymerase. Science. 1969; 166:393–395. - PubMed
-
- Raal F.J., Santos R.D., Blom D.J., Marais A.D., Charng M.-J., Cromwell W.C., Lachmann R.H., Gaudet D., Tan J.L., Chasan-Taber S. et al. . Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010; 375:998–1006. - PubMed
-
- Bennett C.F., Swayze E.E.. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010; 50:259–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

