Peripheral blood gene expression reveals an inflammatory transcriptomic signature in Friedreich's ataxia patients
- PMID: 29790959
- PMCID: PMC6097013
- DOI: 10.1093/hmg/ddy198
Peripheral blood gene expression reveals an inflammatory transcriptomic signature in Friedreich's ataxia patients
Abstract
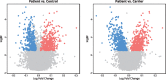
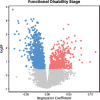
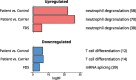
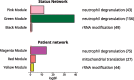
Transcriptional changes in Friedreich's ataxia (FRDA), a rare and debilitating recessive Mendelian neurodegenerative disorder, have been studied in affected but inaccessible tissues-such as dorsal root ganglia, sensory neurons and cerebellum-in animal models or small patient series. However, transcriptional changes induced by FRDA in peripheral blood, a readily accessible tissue, have not been characterized in a large sample. We used differential expression, association with disability stage, network analysis and enrichment analysis to characterize the peripheral blood transcriptome and identify genes that were differentially expressed in FRDA patients (n = 418) compared with both heterozygous expansion carriers (n = 228) and controls (n = 93 739 individuals in total), or were associated with disease progression, resulting in a disease signature for FRDA. We identified a transcriptional signature strongly enriched for an inflammatory innate immune response. Future studies should seek to further characterize the role of peripheral inflammation in FRDA pathology and determine its relevance to overall disease progression.
Figures




References
-
- Cossée M., Puccio H., Gansmuller A., Koutnikova H., Dierich A., LeMeur M.. et al. (2000) Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum. Mol. Genet., 9, 1219–1226. - PubMed
-
- Pastore A., Puccio H. (2013) Frataxin: a protein in search for a function. J. Neurochem., 126, 43–52. - PubMed
-
- Cnop M., Mulder H., Igoillo-Esteve M. (2013) Diabetes in Friedreich ataxia. J. Neurochem., 126, 94–102. - PubMed
-
- Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A.. et al. (1996) Triplet repeat expansion Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science, 271, 1423–1427. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

