The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update
- PMID: 29790989
- PMCID: PMC6030816
- DOI: 10.1093/nar/gky379
The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update
Abstract
Galaxy (homepage: https://galaxyproject.org, main public server: https://usegalaxy.org) is a web-based scientific analysis platform used by tens of thousands of scientists across the world to analyze large biomedical datasets such as those found in genomics, proteomics, metabolomics and imaging. Started in 2005, Galaxy continues to focus on three key challenges of data-driven biomedical science: making analyses accessible to all researchers, ensuring analyses are completely reproducible, and making it simple to communicate analyses so that they can be reused and extended. During the last two years, the Galaxy team and the open-source community around Galaxy have made substantial improvements to Galaxy's core framework, user interface, tools, and training materials. Framework and user interface improvements now enable Galaxy to be used for analyzing tens of thousands of datasets, and >5500 tools are now available from the Galaxy ToolShed. The Galaxy community has led an effort to create numerous high-quality tutorials focused on common types of genomic analyses. The Galaxy developer and user communities continue to grow and be integral to Galaxy's development. The number of Galaxy public servers, developers contributing to the Galaxy framework and its tools, and users of the main Galaxy server have all increased substantially.
Figures
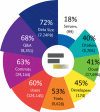
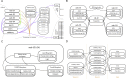

References
-
- Yoo A.B., Jette M.A., Grondona M.. SLURM: Simple Linux Utility for Resource Management. Job Scheduling Strategies for Parallel Processing, Lecture Notes in Computer Science. 2003; Berlin, Heidelberg: Springer; 44–60.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

