A genome-wide microRNA screen identifies regulators of tetraploid cell proliferation
- PMID: 29791254
- PMCID: PMC6080710
- DOI: 10.1091/mbc.E18-02-0141
A genome-wide microRNA screen identifies regulators of tetraploid cell proliferation
Abstract
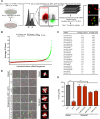
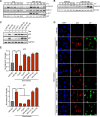
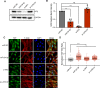
Tetraploid cells, which are most commonly generated by errors in cell division, are genomically unstable and have been shown to promote tumorigenesis. Recent genomic studies have estimated that ∼40% of all solid tumors have undergone a genome-doubling event during their evolution, suggesting a significant role for tetraploidy in driving the development of human cancers. To safeguard against the deleterious effects of tetraploidy, nontransformed cells that fail mitosis and become tetraploid activate both the Hippo and p53 tumor suppressor pathways to restrain further proliferation. Tetraploid cells must therefore overcome these antiproliferative barriers to ultimately drive tumor development. However, the genetic routes through which spontaneously arising tetraploid cells adapt to regain proliferative capacity remain poorly characterized. Here, we conducted a comprehensive gain-of-function genome-wide screen to identify microRNAs (miRNAs) that are sufficient to promote the proliferation of tetraploid cells. Our screen identified 23 miRNAs whose overexpression significantly promotes tetraploid proliferation. The vast majority of these miRNAs facilitate tetraploid growth by enhancing mitogenic signaling pathways (e.g., miR-191-3p); however, we also identified several miRNAs that impair the p53/p21 pathway (e.g., miR-523-3p), and a single miRNA (miR-24-3p) that potently inactivates the Hippo pathway via down-regulation of the tumor suppressor gene NF2. Collectively, our data reveal several avenues through which tetraploid cells may regain the proliferative capacity necessary to drive tumorigenesis.
Figures

References
-
- Carter SB. (1967). Effects of cytochalasins on mammalian cells. Nature , 261–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

