Tracking of Engineered Bacteria In Vivo Using Nonstandard Amino Acid Incorporation
- PMID: 29791796
- PMCID: PMC6415965
- DOI: 10.1021/acssynbio.8b00135
Tracking of Engineered Bacteria In Vivo Using Nonstandard Amino Acid Incorporation
Abstract
The rapidly growing field of microbiome research presents a need for better methods of monitoring gut microbes in vivo with high spatial and temporal resolution. We report a method of tracking microbes in vivo within the gastrointestinal tract by programming them to incorporate nonstandard amino acids (NSAA) and labeling them via click chemistry. Using established machinery constituting an orthogonal translation system (OTS), we engineered Escherichia coli to incorporate p-azido-l-phenylalanine (pAzF) in place of the UAG (amber) stop codon. We also introduced a mutant gene encoding for a cell surface protein (CsgA) that was altered to contain an in-frame UAG codon. After pAzF incorporation and extracellular display, the engineered strains could be covalently labeled via copper-free click reaction with a Cy5 dye conjugated to the dibenzocyclooctyl (DBCO) group. We confirmed the functionality of the labeling strategy in vivo using a murine model. Labeling of the engineered strain could be observed using oral administration of the dye to mice several days after colonization of the gastrointestinal tract. This work sets the foundation for the development of in vivo tracking microbial strategies that may be compatible with noninvasive imaging modalities and are capable of longitudinal spatiotemporal monitoring of specific microbial populations.
Keywords: click chemistry; curli fibers; microbiome imaging; nonstandard amino acid.
Conflict of interest statement
The authors have no competing financial interests to report.
Figures

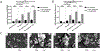

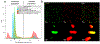

References
-
- Lynch SV, and Pedersen O (2016) The Human Intestinal Microbiome in Health and Disease, New England Journal ofMedicine 375, 2369–2379. - PubMed
-
- Thomas V, Clark J, and Doré J (2015) Fecal microbiota analysis: an overview of sample collection methods and sequencing strategies, Future Microbiology 10, 1485–1504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

