De novo transcriptome assembly and positive selection analysis of an individual deep-sea fish
- PMID: 29793428
- PMCID: PMC5968573
- DOI: 10.1186/s12864-018-4720-z
De novo transcriptome assembly and positive selection analysis of an individual deep-sea fish
Abstract
Background: High hydrostatic pressure and low temperatures make the deep sea a harsh environment for life forms. Actin organization and microtubules assembly, which are essential for intracellular transport and cell motility, can be disrupted by high hydrostatic pressure. High hydrostatic pressure can also damage DNA. Nucleic acids exposed to low temperatures can form secondary structures that hinder genetic information processing. To study how deep-sea creatures adapt to such a hostile environment, one of the most straightforward ways is to sequence and compare their genes with those of their shallow-water relatives.
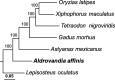
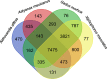
Results: We captured an individual of the fish species Aldrovandia affinis, which is a typical deep-sea inhabitant, from the Okinawa Trough at a depth of 1550 m using a remotely operated vehicle (ROV). We sequenced its transcriptome and analyzed its molecular adaptation. We obtained 27,633 protein coding sequences using an Illumina platform and compared them with those of several shallow-water fish species. Analysis of 4918 single-copy orthologs identified 138 positively selected genes in A. affinis, including genes involved in microtubule regulation. Particularly, functional domains related to cold shock as well as DNA repair are exposed to positive selection pressure in both deep-sea fish and hadal amphipod.
Conclusions: Overall, we have identified a set of positively selected genes related to cytoskeleton structures, DNA repair and genetic information processing, which shed light on molecular adaptation to the deep sea. These results suggest that amino acid substitutions of these positively selected genes may contribute crucially to the adaptation of deep-sea animals. Additionally, we provide a high-quality transcriptome of a deep-sea fish for future deep-sea studies.
Keywords: Cold shock; Cytoskeleton; High hydrostatic pressure; Microtubule; Positive selection.
Conflict of interest statement
Ethics approval and consent to participate
All animal experiments were approved by the Ethics Committee of the Hong Kong University of Science and Technology.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

