Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus
- PMID: 29793506
- PMCID: PMC5966903
- DOI: 10.1186/s12929-018-0449-x
Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus
Abstract
Background: Middle East respiratory syndrome coronavirus (MERS-CoV) consists of a positive-sense, single-stranded RNA genome and four structural proteins: the spike, envelope, membrane, and nucleocapsid protein. The assembly of the viral genome into virus particles involves viral structural proteins and is believed to be mediated through recognition of specific sequences and RNA structures of the viral genome.
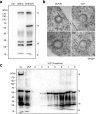
Methods and results: A culture system for the production of MERS coronavirus-like particles (MERS VLPs) was determined and established by electron microscopy and the detection of coexpressed viral structural proteins. Using the VLP system, a 258-nucleotide RNA fragment, which spans nucleotides 19,712 to 19,969 of the MERS-CoV genome (designated PS258(19712-19969)ME), was identified to function as a packaging signal. Assembly of the RNA packaging signal into MERS VLPs is dependent on the viral nucleocapsid protein. In addition, a 45-nucleotide stable stem-loop substructure of the PS258(19712-19969)ME interacted with both the N-terminal domain and the C-terminal domain of the viral nucleocapsid protein. Furthermore, a functional SARS-CoV RNA packaging signal failed to assemble into the MERS VLPs, which indicated virus-specific assembly of the RNA genome.
Conclusions: A MERS-oV RNA packaging signal was identified by the detection of GFP expression following an incubation of MERS VLPs carrying the heterologous mRNA GFP-PS258(19712-19969)ME with virus permissive Huh7 cells. The MERS VLP system could help us in understanding virus infection and morphogenesis.
Keywords: MERS-CoV; Nucleocapsid protein; RNA packaging signal.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent.J Virol. 2005 Nov;79(22):13848-55. doi: 10.1128/JVI.79.22.13848-13855.2005. J Virol. 2005. PMID: 16254320 Free PMC article.
-
The Endonucleolytic RNA Cleavage Function of nsp1 of Middle East Respiratory Syndrome Coronavirus Promotes the Production of Infectious Virus Particles in Specific Human Cell Lines.J Virol. 2018 Oct 12;92(21):e01157-18. doi: 10.1128/JVI.01157-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111568 Free PMC article.
-
Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication.mBio. 2017 Nov 21;8(6):e01658-17. doi: 10.1128/mBio.01658-17. mBio. 2017. PMID: 29162711 Free PMC article.
-
Coronavirus genomic RNA packaging.Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review.
-
The SARS coronavirus nucleocapsid protein--forms and functions.Antiviral Res. 2014 Mar;103:39-50. doi: 10.1016/j.antiviral.2013.12.009. Epub 2014 Jan 11. Antiviral Res. 2014. PMID: 24418573 Free PMC article. Review.
Cited by
-
Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.FEBS J. 2022 Jul;289(13):3813-3825. doi: 10.1111/febs.16239. Epub 2021 Oct 30. FEBS J. 2022. PMID: 34665939 Free PMC article.
-
Sequential glycosylations at the multibasic cleavage site of SARS-CoV-2 spike protein regulate viral activity.Nat Commun. 2024 May 16;15(1):4162. doi: 10.1038/s41467-024-48503-x. Nat Commun. 2024. PMID: 38755139 Free PMC article.
-
One Year of SARS-CoV-2: How Much Has the Virus Changed?Biology (Basel). 2021 Jan 26;10(2):91. doi: 10.3390/biology10020091. Biology (Basel). 2021. PMID: 33530355 Free PMC article.
-
Control Measures for SARS-CoV-2: A Review on Light-Based Inactivation of Single-Stranded RNA Viruses.Pathogens. 2020 Sep 8;9(9):737. doi: 10.3390/pathogens9090737. Pathogens. 2020. PMID: 32911671 Free PMC article. Review.
-
A synthetic defective interfering SARS-CoV-2.PeerJ. 2021 Jul 1;9:e11686. doi: 10.7717/peerj.11686. eCollection 2021. PeerJ. 2021. PMID: 34249513 Free PMC article.
References
-
- Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

