Maternal omega-3 fatty acids regulate offspring obesity through persistent modulation of gut microbiota
- PMID: 29793531
- PMCID: PMC5968592
- DOI: 10.1186/s40168-018-0476-6
Maternal omega-3 fatty acids regulate offspring obesity through persistent modulation of gut microbiota
Abstract
Background: The early-life gut microbiota plays a critical role in host metabolism in later life. However, little is known about how the fatty acid profile of the maternal diet during gestation and lactation influences the development of the offspring gut microbiota and subsequent metabolic health outcomes.
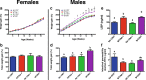
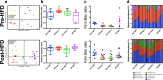
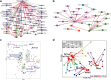
Results: Here, using a unique transgenic model, we report that maternal endogenous n-3 polyunsaturated fatty acid (PUFA) production during gestation or lactation significantly reduces weight gain and markers of metabolic disruption in male murine offspring fed a high-fat diet. However, maternal fatty acid status appeared to have no significant effect on weight gain in female offspring. The metabolic phenotypes in male offspring appeared to be mediated by comprehensive restructuring of gut microbiota composition. Reduced maternal n-3 PUFA exposure led to significantly depleted Epsilonproteobacteria, Bacteroides, and Akkermansia and higher relative abundance of Clostridia. Interestingly, offspring metabolism and microbiota composition were more profoundly influenced by the maternal fatty acid profile during lactation than in utero. Furthermore, the maternal fatty acid profile appeared to have a long-lasting effect on offspring microbiota composition and function that persisted into adulthood after life-long high-fat diet feeding.
Conclusions: Our data provide novel evidence that weight gain and metabolic dysfunction in adulthood is mediated by maternal fatty acid status through long-lasting restructuring of the gut microbiota. These results have important implications for understanding the interaction between modern Western diets, metabolic health, and the intestinal microbiome.
Keywords: Maternal diet; Microbiome; Microbiota; Obesity; n-3 PUFA.
Conflict of interest statement
Ethics approval
All animal procedures in this study were performed in accordance with the ethical guidelines approved by the MGH Subcommittee on Research Animal Care.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Omega-3 polyunsaturated fatty acids critically regulate behaviour and gut microbiota development in adolescence and adulthood.Brain Behav Immun. 2017 Jan;59:21-37. doi: 10.1016/j.bbi.2016.07.145. Epub 2016 Jul 14. Brain Behav Immun. 2017. PMID: 27423492
-
Maternal n-3 polyunsaturated fatty acids restructure gut microbiota of offspring mice and decrease their susceptibility to mammary gland cancer.Food Funct. 2021 Sep 7;12(17):8154-8168. doi: 10.1039/d1fo00906k. Epub 2021 Jul 22. Food Funct. 2021. PMID: 34291263
-
The Transplantation of ω3 PUFA-Altered Gut Microbiota of fat-1 Mice to Wild-Type Littermates Prevents Obesity and Associated Metabolic Disorders.Diabetes. 2018 Aug;67(8):1512-1523. doi: 10.2337/db17-1488. Epub 2018 May 23. Diabetes. 2018. PMID: 29793999
-
Impact of dietary fat on gut microbiota and low-grade systemic inflammation: mechanisms and clinical implications on obesity.Int J Food Sci Nutr. 2018 Mar;69(2):125-143. doi: 10.1080/09637486.2017.1343286. Epub 2017 Jul 4. Int J Food Sci Nutr. 2018. PMID: 28675945 Review.
-
N-3 polyunsaturated fatty acids: An innovative strategy against obesity and related metabolic disorders, intestinal alteration and gut microbiota dysbiosis.Biochimie. 2019 Apr;159:66-71. doi: 10.1016/j.biochi.2019.01.017. Epub 2019 Jan 25. Biochimie. 2019. PMID: 30690133 Review.
Cited by
-
Decreased Tissue Omega-6/Omega-3 Fatty Acid Ratio Prevents Chemotherapy-Induced Gastrointestinal Toxicity Associated with Alterations of Gut Microbiome.Int J Mol Sci. 2022 May 10;23(10):5332. doi: 10.3390/ijms23105332. Int J Mol Sci. 2022. PMID: 35628140 Free PMC article.
-
Plasma Levels of Omega-3 and Omega-6 Derived Oxylipins Are Associated with Fecal Microbiota Composition in Young Adults.Nutrients. 2022 Nov 24;14(23):4991. doi: 10.3390/nu14234991. Nutrients. 2022. PMID: 36501021 Free PMC article.
-
Postnatal supplementation with alarmins S100a8/a9 ameliorates malnutrition-induced neonate enteropathy in mice.Nat Commun. 2024 Oct 4;15(1):8623. doi: 10.1038/s41467-024-52829-x. Nat Commun. 2024. PMID: 39366940 Free PMC article.
-
Maternal Plasma Glycerophospholipids LC-PUFA Levels Have a Sex-Specific Association with the Offspring's Cord Plasma Glycerophospholipids-Fatty Acid Desaturation Indices at Birth.Int J Environ Res Public Health. 2022 Nov 11;19(22):14850. doi: 10.3390/ijerph192214850. Int J Environ Res Public Health. 2022. PMID: 36429569 Free PMC article.
-
Integrated Analyses of Gut Microbiome and Host Metabolome in Children With Henoch-Schönlein Purpura.Front Cell Infect Microbiol. 2022 Jan 25;11:796410. doi: 10.3389/fcimb.2021.796410. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35145922 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

