Restriction of Replication of Oncolytic Herpes Simplex Virus with a Deletion of γ34.5 in Glioblastoma Stem-Like Cells
- PMID: 29793956
- PMCID: PMC6052301
- DOI: 10.1128/JVI.00246-18
Restriction of Replication of Oncolytic Herpes Simplex Virus with a Deletion of γ34.5 in Glioblastoma Stem-Like Cells
Abstract
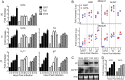
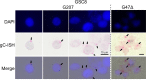
Oncolytic viruses, including herpes simplex viruses (HSVs), are a new class of cancer therapeutic engineered to infect and kill cancer cells while sparing normal tissue. To ensure that oncolytic HSV (oHSV) is safe in the brain, all oHSVs in clinical trial for glioma lack the γ34.5 genes responsible for neurovirulence. However, loss of γ34.5 attenuates growth in cancer cells. Glioblastoma (GBM) is a lethal brain tumor that is heterogeneous and contains a subpopulation of cancer stem cells, termed GBM stem-like cells (GSCs), that likely promote tumor progression and recurrence. GSCs and matched serum-cultured GBM cells (ScGCs), representative of bulk or differentiated tumor cells, were isolated from the same patient tumor specimens. ScGCs are permissive to replication and cell killing by oHSV with deletion of the γ34.5 genes (γ34.5- oHSV), while patient-matched GSCs were not, implying an underlying biological difference between stem and bulk cancer cells. GSCs specifically restrict the synthesis of HSV-1 true late (TL) proteins, without affecting viral DNA replication or transcription of TL genes. A global shutoff of cellular protein synthesis also occurs late after γ34.5- oHSV infection of GSCs but does not affect the synthesis of early and leaky late viral proteins. Levels of phosphorylated eIF2α and eIF4E do not correlate with cell permissivity. Expression of Us11 in GSCs rescues replication of γ34.5- oHSV. The difference in degrees of permissivity between GSCs and ScGCs to γ34.5- oHSV illustrates a selective translational regulatory pathway in GSCs that may be operative in other stem-like cells and has implications for creating oHSVs.IMPORTANCE Herpes simplex virus (HSV) can be genetically engineered to endow cancer-selective replication and oncolytic activity. γ34.5, a key neurovirulence gene, has been deleted in all oncolytic HSVs in clinical trial for glioma. Glioblastoma stem-like cells (GSCs) are a subpopulation of tumor cells thought to drive tumor heterogeneity and therapeutic resistance. GSCs are nonpermissive for γ34.5- HSV, while non-stem-like cancer cells from the same patient tumors are permissive. GSCs restrict true late protein synthesis, despite normal viral DNA replication and transcription of all kinetic classes. This is specific for true late translation as early and leaky late transcripts are translated late in infection, notwithstanding shutoff of cellular protein synthesis. Expression of Us11 in GSCs rescues the replication of γ34.5- HSV. We have identified a cell type-specific innate response to HSV-1 that limits oncolytic activity in glioblastoma.
Keywords: HSV; Us11; cancer stem cell; glioma; herpes simplex virus; oncolytic virus; translation.
Copyright © 2018 American Society for Microbiology.
Figures



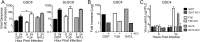



References
-
- Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. 2017. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol 19(Supple 5):v1–v88. doi: 10.1093/neuonc/nox158. - DOI - PMC - PubMed
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. doi: 10.1056/NEJMoa043330. - DOI - PubMed
-
- Lan X, Jorg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, Park NI, Coutinho FJ, Whetstone H, Selvadurai HJ, Che C, Luu B, Carles A, Moksa M, Rastegar N, Head R, Dolma S, Prinos P, Cusimano MD, Das S, Bernstein M, Arrowsmith CH, Mungall AJ, Moore RA, Ma Y, Gallo M, Lupien M, Pugh TJ, Taylor MD, Hirst M, Eaves CJ, Simons BD, Dirks PB. 2017. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549:227–232. doi: 10.1038/nature23666. - DOI - PMC - PubMed
-
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. doi: 10.1126/science.1254257. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

