The Extra-Pathway Interactome of the TCA Cycle: Expected and Unexpected Metabolic Interactions
- PMID: 29794018
- PMCID: PMC6052981
- DOI: 10.1104/pp.17.01687
The Extra-Pathway Interactome of the TCA Cycle: Expected and Unexpected Metabolic Interactions
Abstract
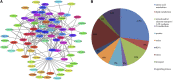
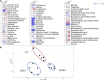
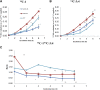
The plant tricarboxylic acid (TCA) cycle provides essential precursors for respiration, amino acid biosynthesis, and general nitrogen metabolism; moreover, it is closely involved in biotic stress responses and cellular redox homeostasis. To further understand the in vivo function of the TCA cycle enzymes, we combined affinity purification with proteomics to generate a comprehensive extra-pathway protein-protein interaction network of the plant TCA cycle. We identified 125 extra-pathway interactions in Arabidopsis (Arabidopsis thaliana) mostly related to the mitochondrial electron transport complex/ATP synthesis and amino acid metabolism but also to proteins associated with redox stress. We chose three high-scoring and two low-scoring interactions for complementary bimolecular fluorescence complementation and yeast two-hybrid assays, which highlighted the reliability of our approach, supported the intimate involvement of TCA cycle enzymes within many biological processes, and reflected metabolic changes reported previously for the corresponding mutant lines. To analyze the function of a subset of these interactions, we selected two mutants of mitochondrial glutaredoxin S15 and Amidase, which have not yet been analyzed with respect to their TCA cycle function, and performed metabolite profiling and flux analysis. Consistent with their interactions identified in this study, TCA cycle metabolites and the relative TCA flux of the two mutants were altered significantly.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures

Similar articles
-
Protein-protein interactions and metabolite channelling in the plant tricarboxylic acid cycle.Nat Commun. 2017 May 16;8:15212. doi: 10.1038/ncomms15212. Nat Commun. 2017. PMID: 28508886 Free PMC article.
-
A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism.Plant J. 2011 Aug;67(3):485-98. doi: 10.1111/j.1365-313X.2011.04612.x. Epub 2011 May 31. Plant J. 2011. PMID: 21501262
-
Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):E1392-400. doi: 10.1073/pnas.1424840112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646482 Free PMC article.
-
A kaleidoscopic view of the Arabidopsis core cell cycle interactome.Trends Plant Sci. 2011 Mar;16(3):141-50. doi: 10.1016/j.tplants.2010.12.004. Epub 2011 Jan 11. Trends Plant Sci. 2011. PMID: 21233003 Review.
-
Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport.Curr Opin Plant Biol. 2004 Jun;7(3):254-61. doi: 10.1016/j.pbi.2004.03.007. Curr Opin Plant Biol. 2004. PMID: 15134745 Review.
Cited by
-
Metabolite profiling of Arabidopsis mutants of lower glycolysis.Sci Data. 2022 Oct 11;9(1):614. doi: 10.1038/s41597-022-01673-z. Sci Data. 2022. PMID: 36220829 Free PMC article.
-
Mitochondrial Small Heat Shock Proteins Are Essential for Normal Growth of Arabidopsis thaliana.Front Plant Sci. 2021 Feb 10;12:600426. doi: 10.3389/fpls.2021.600426. eCollection 2021. Front Plant Sci. 2021. PMID: 33643342 Free PMC article.
-
A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts.Nat Commun. 2020 Sep 9;11(1):4509. doi: 10.1038/s41467-020-18234-w. Nat Commun. 2020. PMID: 32908151 Free PMC article.
-
The function of glutaredoxin GRXS15 is required for lipoyl-dependent dehydrogenases in mitochondria.Plant Physiol. 2021 Jul 6;186(3):1507-1525. doi: 10.1093/plphys/kiab172. Plant Physiol. 2021. PMID: 33856472 Free PMC article.
-
Long-term excessive application of K2SO4 fertilizer alters bacterial community and functional pathway of tobacco-planting soil.Front Plant Sci. 2022 Sep 28;13:1005303. doi: 10.3389/fpls.2022.1005303. eCollection 2022. Front Plant Sci. 2022. PMID: 36247599 Free PMC article.
References
-
- Araújo WL, Nunes-Nesi A, Nikoloski Z, Sweetlove LJ, Fernie AR (2012) Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ 35: 1–21 - PubMed
-
- Beeckmans S, Kanarek L (1981) Demonstration of physical interactions between consecutive enzymes of the citric acid cycle and of the aspartate-malate shuttle: a study involving fumarase, malate dehydrogenase, citrate synthesis and aspartate aminotransferase. Eur J Biochem 117: 527–535 - PubMed
-
- Bürckstümmer T, Bennett KL, Preradovic A, Schütze G, Hantschel O, Superti-Furga G, Bauch A (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

