NLR Mutations Suppressing Immune Hybrid Incompatibility and Their Effects on Disease Resistance
- PMID: 29794019
- PMCID: PMC6052992
- DOI: 10.1104/pp.18.00462
NLR Mutations Suppressing Immune Hybrid Incompatibility and Their Effects on Disease Resistance
Abstract
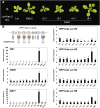
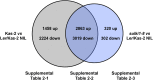
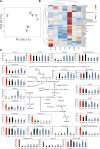
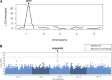
Genetic divergence between populations can lead to reproductive isolation. Hybrid incompatibilities (HI) represent intermediate points along a continuum toward speciation. In plants, genetic variation in disease resistance (R) genes underlies several cases of HI. The progeny of a cross between Arabidopsis (Arabidopsis thaliana) accessions Landsberg erecta (Ler, Poland) and Kashmir2 (Kas2, central Asia) exhibits immune-related HI. This incompatibility is due to a genetic interaction between a cluster of eight TNL (TOLL/INTERLEUKIN1 RECEPTOR-NUCLEOTIDE BINDING-LEU RICH REPEAT) RPP1 (RECOGNITION OF PERONOSPORA PARASITICA1)-like genes (R1-R8) from Ler and central Asian alleles of a Strubbelig-family receptor-like kinase (SRF3) from Kas2. In characterizing mutants altered in Ler/Kas2 HI, we mapped multiple mutations to the RPP1-like Ler locus. Analysis of these suppressor of Ler/Kas2 incompatibility (sulki) mutants reveals complex, additive and epistatic interactions underlying RPP1-like Ler locus activity. The effects of these mutations were measured on basal defense, global gene expression, primary metabolism, and disease resistance to a local Hyaloperonospora arabidopsidis isolate (Hpa Gw) collected from Gorzów (Gw), where the Landsberg accession originated. Gene expression sectors and metabolic hallmarks identified for HI are both dependent and independent of RPP1-like Ler members. We establish that mutations suppressing immune-related Ler/Kas2 HI do not compromise resistance to Hpa Gw. QTL mapping analysis of Hpa Gw resistance point to RPP7 as the causal locus. This work provides insight into the complex genetic architecture of the RPP1-like Ler locus and immune-related HI in Arabidopsis and into the contributions of RPP1-like genes to HI and defense.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures





References
-
- Alcázar R, García AV, Kronholm I, de Meaux J, Koornneef M, Parker JE, Reymond M (2010) Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat Genet 42: 1135–1139 - PubMed
-
- Alcázar R, Pecinka A, Aarts MGM, Fransz PF, Koornneef M (2012) Signals of speciation within Arabidopsis thaliana in comparison with its relatives. Curr Opin Plant Biol 15: 205–211 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

