Discovery of a new Pro-Pro endopeptidase, PPEP-2, provides mechanistic insights into the differences in substrate specificity within the PPEP family
- PMID: 29794027
- PMCID: PMC6052223
- DOI: 10.1074/jbc.RA118.003244
Discovery of a new Pro-Pro endopeptidase, PPEP-2, provides mechanistic insights into the differences in substrate specificity within the PPEP family
Abstract
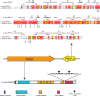
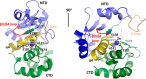
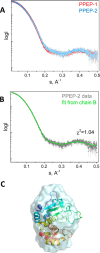
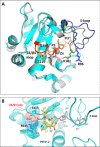
Pro-Pro endopeptidases (PPEPs) belong to a recently discovered family of proteases capable of hydrolyzing a Pro-Pro bond. The first member from the bacterial pathogen Clostridium difficile (PPEP-1) cleaves two C. difficile cell-surface proteins involved in adhesion, one of which is encoded by the gene adjacent to the ppep-1 gene. However, related PPEPs may exist in other bacteria and may shed light on substrate specificity in this enzyme family. Here, we report on the homolog of PPEP-1 in Paenibacillus alvei, which we denoted PPEP-2. We found that PPEP-2 is a secreted metalloprotease, which likewise cleaved a cell-surface protein encoded by an adjacent gene. However, the cleavage motif of PPEP-2, PLP↓PVP, is distinct from that of PPEP-1 (VNP↓PVP). As a result, an optimal substrate peptide for PPEP-2 was not cleaved by PPEP-1 and vice versa. To gain insight into the specificity mechanism of PPEP-2, we determined its crystal structure at 1.75 Å resolution and further confirmed the structure in solution using small-angle X-ray scattering (SAXS). We show that a four-amino-acid loop, which is distinct in PPEP-1 and -2 (GGST in PPEP-1 and SERV in PPEP-2), plays a crucial role in substrate specificity. A PPEP-2 variant, in which the four loop residues had been swapped for those from PPEP-1, displayed a shift in substrate specificity toward PPEP-1 substrates. Our results provide detailed insights into the PPEP-2 structure and the structural determinants of substrate specificity in this new family of PPEP proteases.
Keywords: PPEP; Paenibacillus alvei; Pro-Pro endopeptidase; bacterial adhesion; cell wall; metalloprotease; structural biology; substrate specificity; virulence factor.
© 2018 Klychnikov et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Klein T., Eckhard U., Dufour A., Solis N., and Overall C. M. (2018) Proteolytic cleavage-mechanisms, function, and “Omic” approaches for a near-ubiquitous posttranslational modification. Chem. Rev. 118, 1137–1168 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

