Deep phenotyping in zebrafish reveals genetic and diet-induced adiposity changes that may inform disease risk
- PMID: 29794036
- PMCID: PMC6071777
- DOI: 10.1194/jlr.D084525
Deep phenotyping in zebrafish reveals genetic and diet-induced adiposity changes that may inform disease risk
Abstract
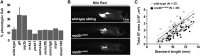
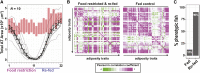
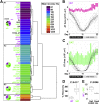
The regional distribution of adipose tissues is implicated in a wide range of diseases. For example, proportional increases in visceral adipose tissue increase the risk for insulin resistance, diabetes, and CVD. Zebrafish offer a tractable model system by which to obtain unbiased and quantitative phenotypic information on regional adiposity, and deep phenotyping can explore complex disease-related adiposity traits. To facilitate deep phenotyping of zebrafish adiposity traits, we used pairwise correlations between 67 adiposity traits to generate stage-specific adiposity profiles that describe changing adiposity patterns and relationships during growth. Linear discriminant analysis classified individual fish according to an adiposity profile with 87.5% accuracy. Deep phenotyping of eight previously uncharacterized zebrafish mutants identified neuropilin 2b as a novel gene that alters adipose distribution. When we applied deep phenotyping to identify changes in adiposity during diet manipulations, zebrafish that underwent food restriction and refeeding had widespread adiposity changes when compared with continuously fed, equivalently sized control animals. In particular, internal adipose tissues (e.g., visceral adipose) exhibited a reduced capacity to replenish lipid following food restriction. Together, these results in zebrafish establish a new deep phenotyping technique as an unbiased and quantitative method to help uncover new relationships between genotype, diet, and adiposity.
Keywords: adipose tissue; fat distribution; obesity.
Copyright © 2018 Minchin et al.
Figures



References
-
- Fox C. S., Massaro J. M., Hoffmann U., Pou K. M., Maurovich-Horvat P., Liu C. Y., Vasan R. S., Murabito J. M., Meigs J. B., Cupples L. A., et al. 2007. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation. 116: 39–48. - PubMed
-
- Karpe F., and Pinnick K. E.. 2015. Biology of upper-body and lower-body adipose tissue–link to whole-body phenotypes. Nat. Rev. Endocrinol. 11: 90–100. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

